Biochemical Characterization and Crystal Structure of a Novel NAD+-Dependent Isocitrate Dehydrogenase from Phaeodactylum tricornutum
Abstract
:1. Introduction
2. Results
2.1. Sequence Analysis
2.2. Overexpression and Purification of PtIDH1
2.3. Kinetics Characterization
2.4. Effects of pH and Temperature
2.5. Effects of Metal Ions and Metabolites
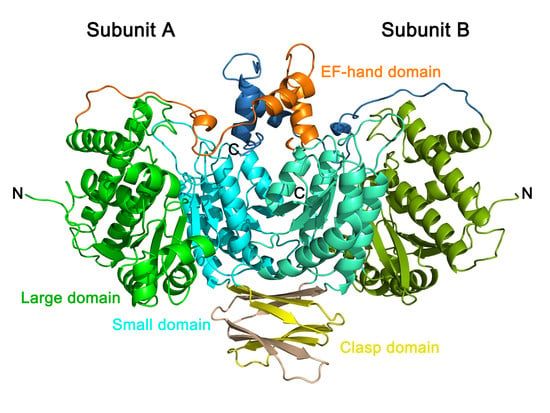
2.6. Crystal Structure of PtIDH1
2.7. Mutational Analysis of PtIDH1
3. Discussion
4. Materials and Methods
4.1. Strains and Cultivation
4.2. Sequence Analysis
4.3. PtIDH1 Gene Cloning and Plasmid Construction
4.4. Site-Directed Mutagenesis
4.5. Recombinant Protein Overexpression and Purification
4.6. Gel Filtration Chromatography
4.7. Circular Dichroism Spectroscopy
4.8. Enzyme Assays and Kinetic Characterization
4.9. Crystallization and Structure Determination
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| PtIDH1 | Phaeodactylum tricornutum isocitrate dehydrogenase 1 |
| NAD+ | β-Nicotinamide adenine dinucleotide |
| NADP+ | β-Nicotinamide-adenine dinucleotide phosphate |
| NAD-IDH | NAD+-dependent isocitrate dehydrogenase |
| NADP-IDH | NADP+-dependent isocitrate dehydrogenase |
| Km | Michaelis constant |
| kcat | catalytic rate constant |
| nH | Hill coefficient |
| CD | circular dichroism |
References
- Chen, R.; Jeong, S.S. Functional prediction: Identification of protein orthologs and paralogs. Protein Sci. 2000, 9, 2344–2353. [Google Scholar] [CrossRef] [Green Version]
- Lin, A.P.; Demeler, B.; Minard, K.I.; Anderson, S.L.; Schirf, V.; Galaleldeen, A.; McAlister-Henn, L. Construction and analyses of tetrameric forms of yeast NAD+-specific isocitrate dehydrogenase. Biochemistry 2011, 50, 230–239. [Google Scholar] [CrossRef] [Green Version]
- Spaans, S.K.; Weusthuis, R.A.; van der Oost, J.; Kengen, S.W. NADPH-generating systems in bacteria and archaea. Front. Microbiol. 2015, 6, 742. [Google Scholar] [CrossRef]
- Leterrier, M.; Barroso, J.B.; Valderrama, R.; Palma, J.M.; Corpas, F.J. NADP-dependent isocitrate dehydrogenase from Arabidopsis roots contributes in the mechanism of defence against the nitro-oxidative stress induced by salinity. Sci. World J. 2012, 2012, 694740. [Google Scholar] [CrossRef] [Green Version]
- Mhamdi, A.; Mauve, C.; Gouia, H.; Saindrenan, P.; Hodges, M.; Noctor, G. Cytosolic NADP-dependent isocitrate dehydrogenase contributes to redox homeostasis and the regulation of pathogen responses in Arabidopsis leaves. Plant Cell Environ. 2010, 33, 1112–1123. [Google Scholar] [PubMed]
- Leterrier, M.; Del Rio, L.A.; Corpas, F.J. Cytosolic NADP-isocitrate dehydrogenase of pea plants: Genomic clone characterization and functional analysis under abiotic stress conditions. Free Radic. Res. 2007, 41, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Waitkus, M.S.; Diplas, B.H.; Yan, H. Biological role and therapeutic potential of IDH mutations in cancer. Cancer Cell 2018, 34, 186–195. [Google Scholar] [CrossRef] [Green Version]
- Molenaar, R.J.; Maciejewski, J.P.; Wilmink, J.W.; van Noorden, C.J.F. Wild-type and mutated IDH1/2 enzymes and therapy responses. Oncogene 2018, 37, 1949–1960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waitkus, M.S.; Diplas, B.H.; Yan, H. Isocitrate dehydrogenase mutations in gliomas. Neuro-Oncology. 2016, 18, 16–26. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Wu, Y.; Liu, J.; Song, P.; Li, S.; Zhou, X.; Zhu, G. Crystal structure of the isocitrate dehydrogenase 2 from Acinetobacter baumannii (AbIDH2) reveals a novel dimeric structure with two monomeric IDH-like subunits. Int. J. Mol. Sci. 2018, 19, 1131. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Lv, C.; Zhu, G. Novel type II and monomeric NAD+ specific isocitrate dehydrogenases: Phylogenetic affinity, enzymatic characterization, and evolutionary implication. Sci. Rep. 2015, 5, 9150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, W.G.; Song, P.; Cao, Z.Y.; Wang, P.; Zhu, G.P. A unique homodimeric NAD+-linked isocitrate dehydrogenase from the smallest autotrophic eukaryote Ostreococcus tauri. FASEB J. 2015, 29, 2462–2472. [Google Scholar] [CrossRef] [Green Version]
- Dean, A.M.; Koshland, D.E., Jr. Kinetic mechanism of Escherichia coli isocitrate dehydrogenase. Biochemistry 1993, 32, 9302–9309. [Google Scholar] [CrossRef]
- Chen, R.; Greer, A.F.; Dean, A.M. Structural constraints in protein engineering. The coenzyme specificity of Escherichia coli isocitrate dehydrogenase. Eur. J. Biochem. 1997, 250, 578–582. [Google Scholar] [CrossRef]
- Wang, S.; Shen, Q.; Chen, G.; Zheng, J.; Tan, H.; Jia, Z. The phosphatase mechanism of bifunctional kinase/phosphatase AceK. Chem. Commun. (Camb.) 2014, 50, 14117–14120. [Google Scholar] [CrossRef]
- Xu, X.; Zhao, J.; Xu, Z.; Peng, B.; Huang, Q.; Arnold, E.; Ding, J. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J. Biol. Chem. 2004, 279, 33946–33957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imada, K.; Tamura, T.; Takenaka, R.; Kobayashi, I.; Namba, K.; Inagaki, K. Structure and quantum chemical analysis of NAD+-dependent isocitrate dehydrogenase: Hydride transfer and co-factor specificity. Proteins 2008, 70, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Zhong, C.; Huang, W.; Ding, J. Structural studies of Saccharomyces cerevesiae mitochondrial NADP-dependent isocitrate dehydrogenase in different enzymatic states reveal substantial conformational changes during the catalytic reaction. Protein Sci. 2008, 17, 1542–1554. [Google Scholar] [CrossRef] [Green Version]
- Ceccarelli, C.; Grodsky, N.B.; Ariyaratne, N.; Colman, R.F.; Bahnson, B.J. Crystal structure of porcine mitochondrial NADP+-dependent isocitrate dehydrogenase complexed with Mn2+ and isocitrate. Insights into the enzyme mechanism. J. Biol. Chem. 2002, 277, 43454–43462. [Google Scholar] [CrossRef] [Green Version]
- Zhu, B.H.; Zhang, R.H.; Lv, N.N.; Yang, G.P.; Wang, Y.S.; Pan, K.H. The role of malic enzyme on promoting total lipid and fatty acid production in Phaeodactylum tricornutum. Front. Plant Sci. 2018, 9, 826. [Google Scholar] [CrossRef]
- Xue, J.; Balamurugan, S.; Li, D.W.; Liu, Y.H.; Zeng, H.; Wang, L.; Yang, W.D.; Liu, J.S.; Li, H.Y. Glucose-6-phosphate dehydrogenase as a target for highly efficient fatty acid biosynthesis in microalgae by enhancing NADPH supply. Metab. Eng. 2017, 41, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Bowler, C.; Allen, A.E.; Badger, J.H.; Grimwood, J.; Jabbari, K.; Kuo, A.; Maheswari, U.; Martens, C.; Maumus, F.; Otillar, R.P.; et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 2008, 456, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Sabir, J.S.M.; Theriot, E.C.; Manning, S.R.; Al-Malki, A.L.; Khiyami, M.A.; Al-Ghamdi, A.K.; Sabir, M.J.; Romanovicz, D.K.; Hajrah, N.H.; El Omri, A.; et al. Phylogenetic analysis and a review of the history of the accidental phytoplankter, Phaeodactylum tricornutum Bohlin (Bacillariophyta). PLoS ONE 2018, 13, e0196744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanc-Mathieu, R.; Verhelst, B.; Derelle, E.; Rombauts, S.; Bouget, F.Y.; Carre, I.; Chateau, A.; Eyre-Walker, A.; Grimsley, N.; Moreau, H.; et al. An improved genome of the model marine alga Ostreococcus tauri unfolds by assessing Illumina de novo assemblies. BMC Genom. 2014, 15, 1103. [Google Scholar] [CrossRef] [Green Version]
- Daubin, V.; Szollosi, G.J. Horizontal Gene Transfer and the History of Life. Cold Spring Harb. Perspect. Biol. 2016, 8, a018036. [Google Scholar] [CrossRef] [Green Version]
- Steen, I.H.; Madern, D.; Karlstrom, M.; Lien, T.; Ladenstein, R.; Birkeland, N.K. Comparison of isocitrate dehydrogenase from three hyperthermophiles reveals differences in thermostability, cofactor specificity, oligomeric state, and phylogenetic affiliation. J. Biol. Chem. 2001, 276, 43924–43931. [Google Scholar] [CrossRef] [Green Version]
- Karlstrom, M.; Steen, I.H.; Madern, D.; Fedoy, A.E.; Birkeland, N.K.; Ladenstein, R. The crystal structure of a hyperthermostable subfamily II isocitrate dehydrogenase from Thermotoga maritima. FEBS J. 2006, 273, 2851–2868. [Google Scholar] [CrossRef]
- Wu, M.C.; Tian, C.Q.; Cheng, H.M.; Xu, L.; Wang, P.; Zhu, G.P. A novel type II NAD+-specific isocitrate dehydrogenase from the marine bacterium Congregibacter litoralis KT71. PLoS ONE 2015, 10, e0125229. [Google Scholar] [CrossRef]
- Wang, P.; Jin, M.; Zhu, G. Biochemical and molecular characterization of NAD+-dependent isocitrate dehydrogenase from the ethanologenic bacterium Zymomonas mobilis. FEMS Microbiol. Lett. 2012, 327, 134–141. [Google Scholar] [CrossRef] [Green Version]
- Lv, C.; Wang, P.; Wang, W.; Su, R.; Ge, Y.; Zhu, Y.; Zhu, G. Two isocitrate dehydrogenases from a plant pathogen Xanthomonas campestris pv. campestris 8004. Bioinformatic analysis, enzymatic characterization, and implication in virulence. J. Basic Microbiol. 2016, 56, 975–985. [Google Scholar] [CrossRef]
- Lv, P.; Tang, W.; Wang, P.; Cao, Z.; Zhu, G. Enzymatic characterization and functional implication of two structurally different isocitrate dehydrogenases from Xylella fastidiosa. Biotechnol. Appl. Biochem. 2018, 65, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Rivas, J.M.; Vega, J.M. Studies on the isoforms of isocitrate dehydrogenase from Chlamydomonas reinhardtii. J. Plant Physiol. 1994, 143, 129–134. [Google Scholar] [CrossRef]
- Tezuka, T.; Laties, G.G. Isolation and characterization of inner membrane-associated and matrix NAD-specific isocitrate dehydrogenase in potato mitochondria. Plant Physiol. 1983, 72, 959–963. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, P.; Zhu, G.; Wang, B.; Zhu, G. Enzymatic characterization of a type II isocitrate dehydrogenase from pathogenic Leptospira interrogans serovar Lai strain 56601. Appl. Biochem. Biotechnol. 2014, 172, 487–496. [Google Scholar] [CrossRef]
- Huang, S.P.; Cheng, H.M.; Wang, P.; Zhu, G.P. Biochemical characterization and complete conversion of coenzyme specificity of isocitrate dehydrogenase from Bifidobacterium longum. Int. J. Mol. Sci. 2016, 17, 296. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Jin, M.; Su, R.; Song, P.; Wang, M.; Zhu, G. Enzymatic characterization of isocitrate dehydrogenase from an emerging zoonotic pathogen Streptococcus suis. Biochimie 2011, 93, 1470–1475. [Google Scholar] [CrossRef]
- Chen, R.; Yang, H. A highly specific monomeric isocitrate dehydrogenase from Corynebacterium glutamicum. Arch. Biochem. Biophys. 2000, 383, 238–245. [Google Scholar] [CrossRef]
- Wang, A.; Cao, Z.Y.; Wang, P.; Liu, A.M.; Pan, W.; Wang, J.; Zhu, G.P. Heteroexpression and characterization of a monomeric isocitrate dehydrogenase from the multicellular prokaryote Streptomyces avermitilis MA-4680. Mol. Biol. Rep. 2011, 38, 3717–3724. [Google Scholar] [CrossRef]
- Martinez-Rivas, J.M.; Vega, J.M. Purification and characterization of NAD-isocitrate dehydrogenase from Chlamydomonas reinhardtii. Plant Physiol. 1998, 118, 249–255. [Google Scholar] [CrossRef] [Green Version]
- Holm, L.; Rosenstrom, P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010, 38, W545–W549. [Google Scholar] [CrossRef]
- Nelson, M.R.; Thulin, E.; Fagan, P.A.; Forsen, S.; Chazin, W.J. The EF-hand domain: A globally cooperative structural unit. Protein Sci. 2002, 11, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Gifford, J.L.; Walsh, M.P.; Vogel, H.J. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem. J. 2007, 405, 199–221. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.P.; McAlister-Henn, L. Homologous binding sites in yeast isocitrate dehydrogenase for cofactor (NAD+) and allosteric activator (AMP). J. Biol. Chem. 2003, 278, 12864–12872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McIntosh, C.A.; Oliver, D.J. NAD-linked isocitrate dehydrogenase: Isolation, purification, and characterization of the protein from pea mitochondria. Plant Physiol. 1992, 100, 69–75. [Google Scholar] [CrossRef] [Green Version]
- Zhu, G.; Golding, G.B.; Dean, A.M. The selective cause of an ancient adaptation. Science 2005, 307, 1279–1282. [Google Scholar] [CrossRef]
- Steen, I.H.; Lien, T.; Madsen, M.S.; Birkeland, N.K. Identification of cofactor discrimination sites in NAD-isocitrate dehydrogenase from Pyrococcus furiosus. Arch. Microbiol. 2002, 178, 297–300. [Google Scholar] [CrossRef]
- Hurley, J.H.; Chen, R.; Dean, A.M. Determinants of cofactor specificity in isocitrate dehydrogenase: Structure of an engineered NADP+→ NAD+ specificity-reversal mutant. Biochemistry 1996, 35, 5670–5678. [Google Scholar] [CrossRef]
- Kinch, L.N.; Grishin, N.V. Evolution of protein structures and functions. Curr. Opin. Struct. Biol. 2002, 12, 400–408. [Google Scholar] [CrossRef]
- Villalobo, A.; Gonzalez-Munoz, M.; Berchtold, M.W. Proteins with calmodulin-like domains: Structures and functional roles. Cell Mol. Life Sci. 2019, 76, 2299–2328. [Google Scholar] [CrossRef]
- Denton, R.M. Regulation of mitochondrial dehydrogenases by calcium ions. BBA-Bioenergetics 2009, 1787, 1309–1316. [Google Scholar] [CrossRef] [Green Version]
- Yasutake, Y.; Watanabe, S.; Yao, M.; Takada, Y.; Fukunaga, N.; Tanaka, I. Crystal structure of the monomeric isocitrate dehydrogenase in the presence of NADP+: Insight into the cofactor recognition, catalysis, and evolution. J. Biol. Chem. 2003, 278, 36897–36904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimorski, V.; Ku, C.; Martin, W.F.; Gould, S.B. Endosymbiotic theory for organelle origins. Curr. Opin. Microbiol. 2014, 22, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wang, X.; Li, S.; Yao, J.; Shao, Z.; Duan, D. Verification of the Saccharina japonica translocon Tic20 and its localization in the chloroplast membrane in diatoms. Int. J. Mol. Sci. 2019, 20, 4000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sonderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Salvatore, M.; Emanuelsson, O.; Winther, O.; von Heijne, G.; Elofsson, A.; Nielsen, H. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance 2019, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Notredame, C.; Higgins, D.G.; Heringa, J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef] [Green Version]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Bramucci, E.; Paiardini, A.; Bossa, F.; Pascarella, S. PyMod: Sequence similarity searches, multiple sequence-structure alignments, and homology modeling within PyMOL. BMC Bioinformatics 2012, 13 (Suppl. 4), S2. [Google Scholar] [CrossRef] [Green Version]
- Raussens, V.; Ruysschaert, J.M.; Goormaghtigh, E. Protein concentration is not an absolute prerequisite for the determination of secondary structure from circular dichroism spectra: A new scaling method. Anal. Biochem. 2003, 319, 114–121. [Google Scholar] [CrossRef]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, 66 Pt 2, 125–132. [Google Scholar] [CrossRef] [Green Version]
- Hough, M.A.; Wilson, K.S. From crystal to structure with CCP4. Acta Crystallogr. D Biol. Crystallogr. 2018, 74 Pt 2, 67. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60 Pt 12 Pt 1, 2126–2132. [Google Scholar] [CrossRef] [Green Version]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkóczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019, 75 Pt 10, 861–877. [Google Scholar] [CrossRef] [Green Version]
- Laskowski, R.A.; Moss, D.S.; Thornton, J.M. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 1993, 231, 1049–1067. [Google Scholar] [CrossRef] [PubMed]








| Enzyme | NAD+ | NADP+ | ||||
|---|---|---|---|---|---|---|
| Km | kcat | kcat/Km | Km | kcat | kcat/Km | |
| (μM) | (s−1) | (μM−1·s−1) | (μM) | (s−1) | (μM−1·s−1) | |
| PtIDH1 (Mn2+) | 1132.5 ± 23.3 | 180.5 ± 6.8 | 0.16 | − | − | − |
| PtIDH1 (Mg2+) | 903.0 ± 59.9 | 79.0 ± 1.8 | 0.09 | − | − | − |
| O. tauri IDH [12] | 265 | 115 | 0.43 | 4314 | 46 | 0.0107 |
| O. lucimarinus IDH [11] | 136.6 | 60.3 | 0.444 | 2211 | 10 | 0.0045 |
| Micromonas sp. IDH [11] | 126.0 | 22.5 | 0.179 | 1827 | 1.4 | 0.0008 |
| C. litoralis IDH [28] | 309.1 | 84.7 | 0.27 | − | − | − |
| Z. mobilis IDH [29] | 312 | 88 | 0.282 | 8200 | 14 | 0.0017 |
| X. campestris IDH [30] | 225 | 49 | 0.213 | 4322 | 8.5 | 0.002 |
| X. fastidiosa IDH [31] | 121 | 74.6 | 0.617 | 2339 | 6.1 | 0.003 |
| Metal Ions | Relative Activity (%) | Metal Ions | Relative Activity (%) | Metal Ions | Relative Activity (%) |
|---|---|---|---|---|---|
| None | 4.83 ± 0.45 | ||||
| Mn2+ | 100 ± 0.0 | Mn2+ | 100 ± 0.0 | Mg2+ | 100 ± 0.0 |
| Mg2+ | 59.19 ± 3.40 | Mn2+ + Mg2+ | 120.09 ± 9.91 | Mg2+ + Mn2+ | 202.78 ± 3.40 |
| Ca2+ | 3.87 ± 0.43 | Mn2+ + Ca2+ | 107.11 ± 4.00 | Mg2+ + Ca2+ | 58.46 ± 2.52 |
| Co2+ | 5.96 ± 0.39 | Mn2+ + Co2+ | 28.21 ± 1.57 | Mg2+ + Co2+ | 13.48 ± 1.44 |
| Cu2+ | 5.76 ± 0.86 | Mn2+ + Cu2+ | 84.94 ± 6.82 | Mg2+ + Cu2+ | 78.59 ± 1.97 |
| Ni2+ | 1.60 ± 0.36 | Mn2+ + Ni2+ | 70.58 ± 4.47 | Mg2+ + Ni2+ | 28.24 ± 0.85 |
| Na+ | 4.66 ± 0.16 | Mn2+ + Na+ | 114.08 ± 4.93 | Mg2+ + Na+ | 96.87 ± 3.80 |
| Li+ | 5.15 ± 0.30 | Mn2+ + Li+ | 109.11 ± 1.88 | Mg2+ + Li+ | 82.71 ± 5.22 |
| K+ | 4.12 ± 0.52 | Mn2+ + K+ | 99.07 ± 1.61 | Mg2+ + K+ | 97.80 ± 5.82 |
| Metabolite | Concentration (mM) | Relative Activity (%) | ||
|---|---|---|---|---|
| PtIDH1 | O. tauri IDH [12] | C. reinhardtii IDH [39] | ||
| control | 100.00 ± 0.0 | 100 | 100 | |
| citrate | ||||
| 0.01 | 101.98 ± 0.9 | 100.71 | 100 | |
| 0.1 | 101.47 ± 0.2 | 103.54 | 82 | |
| 1.0 | 106.29 ± 0.4 | 106.45 | 64 | |
| α-Ketoglutarate | ||||
| 0.01 | 98.68 ± 1.8 | / | / | |
| 0.1 | 100.69 ± 1.0 | 100.36 | / | |
| 1.0 | 98.29 ± 0.3 | 95.49 | / | |
| ATP | ||||
| 0.01 | 102.06 ± 1.5 | 93.62 | 109 | |
| 0.1 | 98.89 ± 0.4 | 93.62 | 91 | |
| 1.0 | 88.70 ± 0.8 | 81.20 | 54 | |
| ADP | ||||
| 0.01 | 101.04 ± 2.3 | 100.99 | 100 | |
| 0.1 | 97.54 ± 1.5 | 97.87 | 109 | |
| 1.0 | 86.63 ± 0.2 | 95.04 | 91 | |
| AMP | ||||
| 0.01 | 102.02 ± 1.7 | 104.37 | 109 | |
| 0.1 | 99.74 ± 1.0 | 99.07 | 100 | |
| 1.0 | 97.88 ± 4.5 | 89.61 | 91 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, S.-P.; Zhou, L.-C.; Wen, B.; Wang, P.; Zhu, G.-P. Biochemical Characterization and Crystal Structure of a Novel NAD+-Dependent Isocitrate Dehydrogenase from Phaeodactylum tricornutum. Int. J. Mol. Sci. 2020, 21, 5915. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21165915
Huang S-P, Zhou L-C, Wen B, Wang P, Zhu G-P. Biochemical Characterization and Crystal Structure of a Novel NAD+-Dependent Isocitrate Dehydrogenase from Phaeodactylum tricornutum. International Journal of Molecular Sciences. 2020; 21(16):5915. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21165915
Chicago/Turabian StyleHuang, Shi-Ping, Lu-Chun Zhou, Bin Wen, Peng Wang, and Guo-Ping Zhu. 2020. "Biochemical Characterization and Crystal Structure of a Novel NAD+-Dependent Isocitrate Dehydrogenase from Phaeodactylum tricornutum" International Journal of Molecular Sciences 21, no. 16: 5915. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21165915