Amino Acid Deletions in p6Gag Domain of HIV-1 CRF07_BC Ameliorate Galectin-3 Mediated Enhancement in Viral Budding
Abstract
:1. Introduction
2. Results
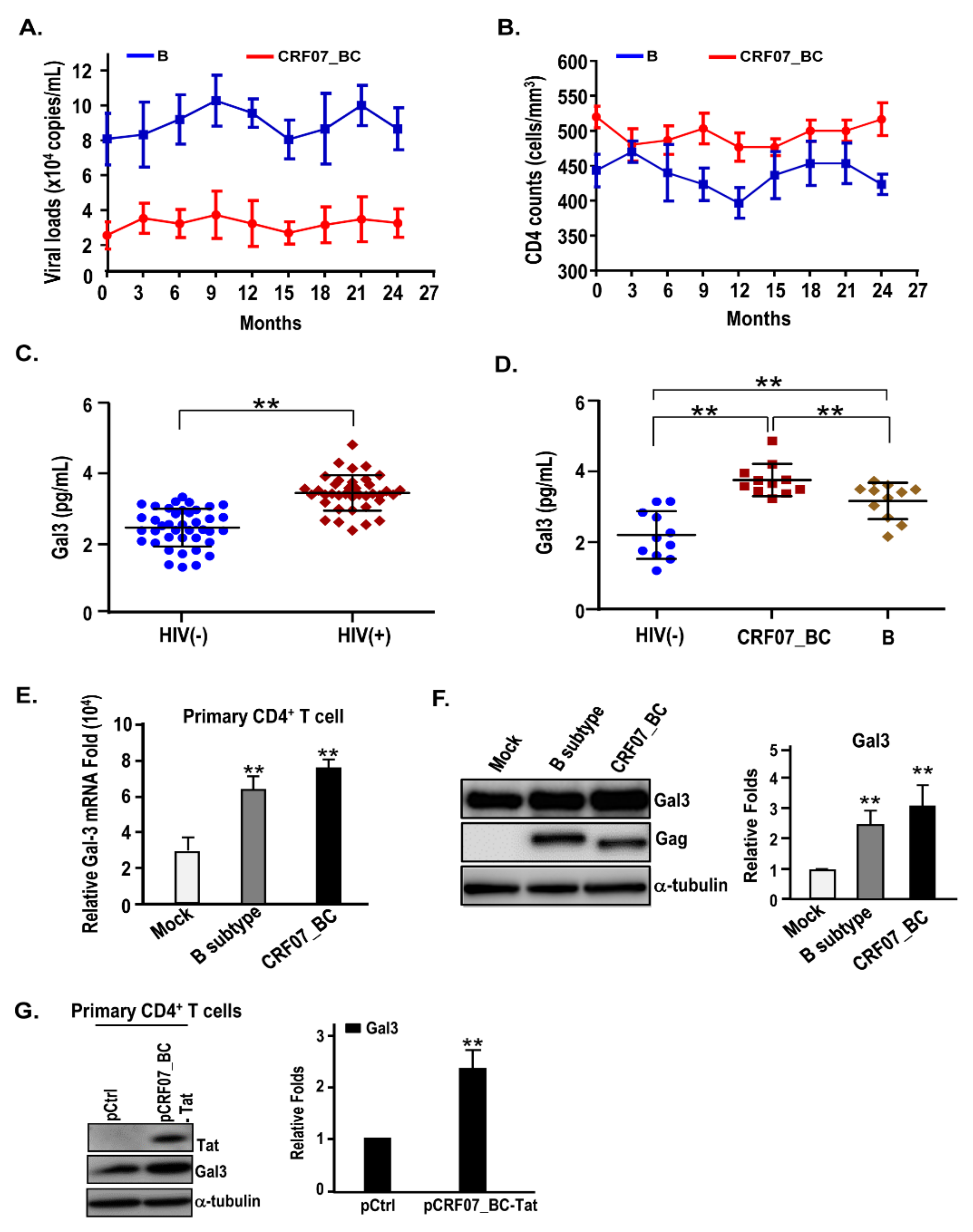
2.1. Slow Disease Progression and Higher Plasma Galectin-3 Detected in HIV-1 CRF07_BC-Infected Patients
2.2. HIV-1 CRF07_BC Infection Induced Galectin-3 Expression
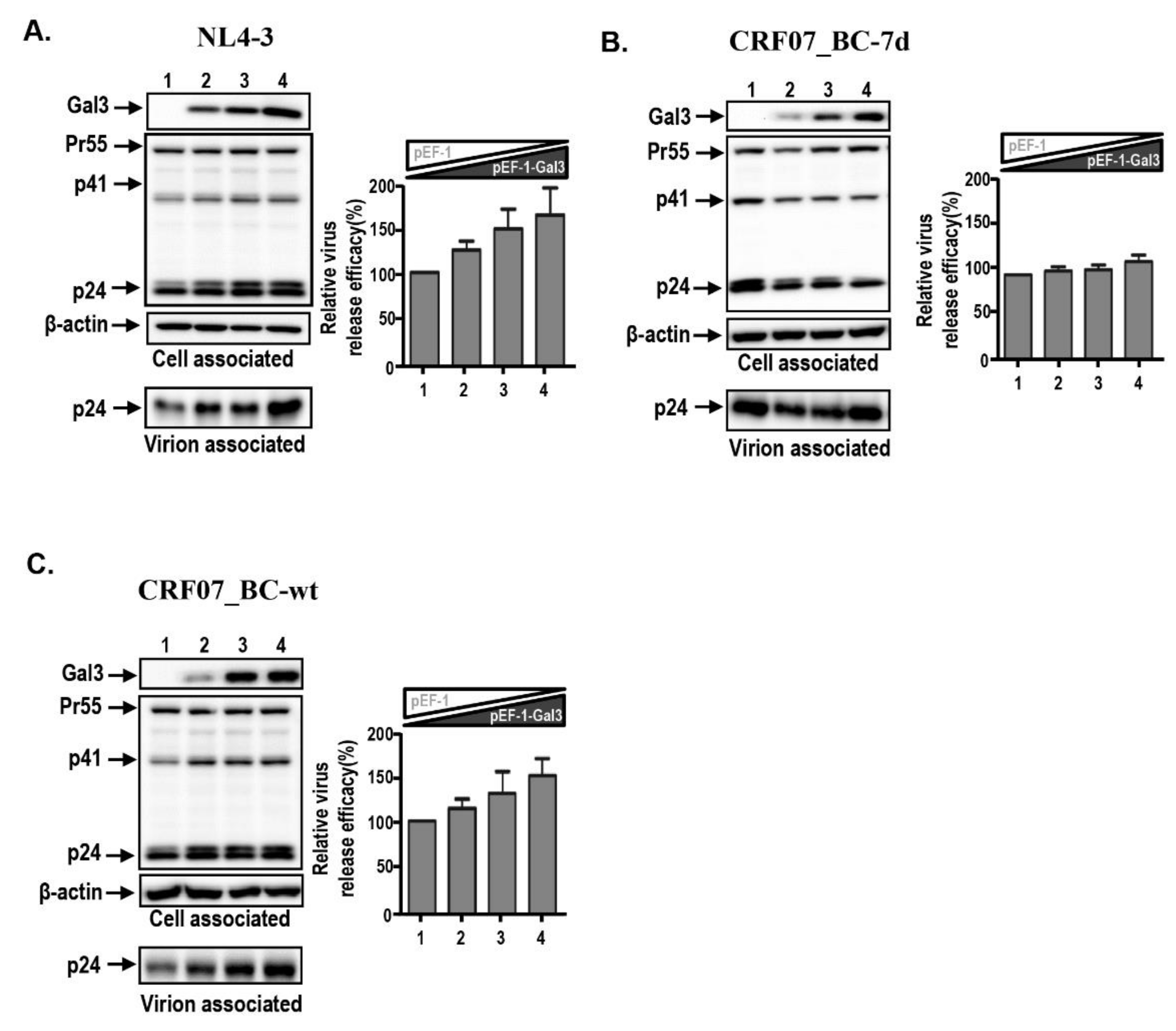
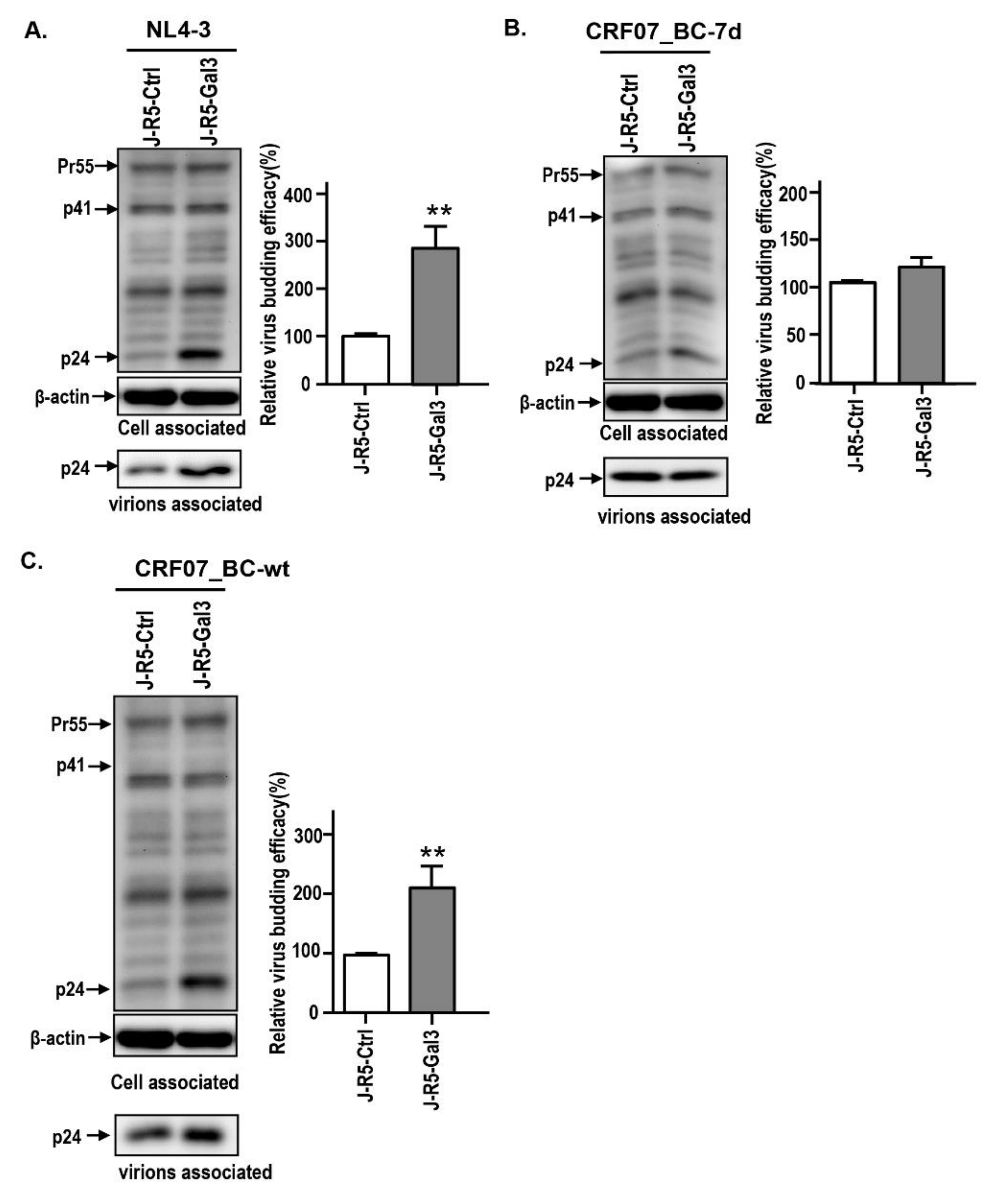
2.3. Amino Acid Deletions in CRF07_BC p6Gag Ameliorated Galectin-3-Mediated Virus Growth and Budding
2.4. Amino Acids Deletions in CRF07_BC p6Gag Attenuated Galectin-3 Dependent on Alix Association with Gag
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Cell and Viruses
4.3. Plasmids
4.4. Determination of HIV-1 Genotypes
4.5. Viral Growth Kinetic Assay
4.6. Immunoblotting
4.7. Co-Immunoprecipitation
4.8. qRT-PCR
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Gal3 | Galectin-3 |
| Alix | ALG-2 interacting protein X |
| CRF07_BC-7d | HIV-1 CRF07_BC virus with 7 amino acid deletions in p6Gag |
| CRF07_BC-wt | HIV-1 CRF07_BC virus non-deletion wild-type |
References
- Su, L.; Graf, M.; Zhang, Y.; von Briesen, H.; Xing, H.; Kostler, J.; Melzl, H.; Wolf, H.; Shao, Y.; Wagner, R. Characterization of a virtually full-length human immunodeficiency virus type 1 genome of a prevalent intersubtype (C/B’) recombinant strain in China. J. Virol. 2000, 74, 11367–11376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, R.; Xia, X.; Kusagawa, S.; Zhang, C.; Ben, K.; Takebe, Y. On-going generation of multiple forms of HIV-1 intersubtype recombinants in the Yunnan Province of China. AIDS 2002, 16, 1401–1407. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.M.; Lan, Y.C.; Lai, S.F.; Yang, J.Y.; Tsai, S.F.; Kuo, S.H. HIV-1 CRF07_BC infections, injecting drug users, Taiwan. Emerg. Infect. Dis. 2006, 12, 703–705. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.Y.; Sheng, W.H.; Lee, C.N.; Sun, H.Y.; Kao, C.L.; Chang, S.F.; Liu, W.C.; Yang, J.Y.; Wong, W.W.; Hung, C.C.; et al. Molecular epidemiology of HIV type 1 subtypes in Taiwan: outbreak of HIV type 1 CRF07_BC infection in intravenous drug users. AIDS Res. Hum. Retrovir. 2006, 22, 1055–1066. [Google Scholar] [CrossRef]
- Lin, H.H.; Shih, Y.L.; Liu, Y.C.; Lee, S.S.; Huang, C.K.; Chen, Y.L.; Chin, C.; Lai, C.H.; Tsai, H.C.; Guo, Y.C.; et al. An epidemic of HIV type I CRF07_BC infection among injection drug users in Taiwan. J. Acquir. Immune. Defic. Syndr. 2006, 42, 248–255. [Google Scholar] [CrossRef]
- Lin, Y.T.; Lan, Y.C.; Chen, Y.J.; Huang, Y.H.; Lee, C.M.; Liu, T.T.; Wong, W.W.; Yang, J.Y.; Wang, C.T.; Chen, Y.M. Molecular epidemiology of HIV-1 infection and full-length genomic analysis of circulating recombinant form 07_BC strains from injection drug users in Taiwan. J. Infect. Dis. 2007, 195, 1283–1293. [Google Scholar] [CrossRef]
- Huang, S.W.; Wang, S.F.; Lin, Y.T.; Yen, C.H.; Lee, C.H.; Wong, W.W.; Tsai, H.C.; Yang, C.J.; Hu, B.S.; Lin, Y.H.; et al. Patients infected with CRF07_BC have significantly lower viral loads than patients with HIV-1 subtype B: mechanism and impact on disease progression. PLoS ONE 2014, 9, e114441. [Google Scholar] [CrossRef]
- Lin, P.H.; Lai, C.C.; Yang, J.L.; Huang, H.L.; Huang, M.S.; Tsai, M.S.; Yang, C.J.; Cheng, C.L.; Su, Y.C.; Chang, S.F.; et al. Slow immunological progression in HIV-1 CRF07_BC-infected injecting drug users. Emerg. Microbes. Infect. 2013, 2, e83. [Google Scholar] [CrossRef]
- Weiss, E.R.; Gottlinger, H. The role of cellular factors in promoting HIV budding. J. Mol. Biol. 2011, 410, 525–533. [Google Scholar] [CrossRef] [Green Version]
- Martin-Serrano, J.; Neil, S.J. Host factors involved in retroviral budding and release. Nat. Rev. Microbiol. 2011, 9, 519–531. [Google Scholar] [CrossRef]
- Jacks, T.; Power, M.D.; Masiarz, F.R.; Luciw, P.A.; Barr, P.J.; Varmus, H.E. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature 1988, 331, 280–283. [Google Scholar] [CrossRef] [PubMed]
- Partin, K.; Krausslich, H.G.; Ehrlich, L.; Wimmer, E.; Carter, C. Mutational analysis of a native substrate of the human immunodeficiency virus type 1 proteinase. J. Virol. 1990, 64, 393847. [Google Scholar] [CrossRef] [Green Version]
- Hurley, J.H.; Emr, S.D. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 2006, 35, 277–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujii, K.; Munshi, U.M.; Ablan, S.D.; Demirov, D.G.; Soheilian, F.; Nagashima, K.; Stephen, A.G.; Fisher, R.J.; Freed, E.O. Functional role of Alix in HIV-1 replication. Virology 2009, 391, 284–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, H.L.; Meng, Z.F.; Zhang, X.Y.; Lu, J.X. [HIV-1 infection affects the expression of host cell factor TSG101 and Alix]. Bing Du Xue Bao 2011, 27, 129–134. [Google Scholar]
- Fujii, K.; Hurley, J.H.; Freed, E.O. Beyond Tsg101: the role of Alix in ‘ESCRTing’ HIV-1. Nat. Rev. Microbiol. 2007, 5, 912–916. [Google Scholar] [CrossRef]
- Liu, F.T.; Bevins, C.L. A sweet target for innate immunity. Nat. Med. 2010, 16, 263–264. [Google Scholar] [CrossRef]
- Vasta, G.R. Roles of galectins in infection. Nat. Rev. Microbiol. 2009, 7, 424–438. [Google Scholar] [CrossRef] [Green Version]
- Diaz-Alvarez, L.; Ortega, E. The Many Roles of Galectin-3, a Multifaceted Molecule, in Innate Immune Responses against Pathogens. Mediat. Inflamm. 2017, 2017, 9247574. [Google Scholar] [CrossRef] [Green Version]
- Thery, C.; Boussac, M.; Veron, P.; Ricciardi-Castagnoli, P.; Raposo, G.; Garin, J.; Amigorena, S. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol. 2001, 166, 7309–7318. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.L.; Chen, Y.H.; Wang, S.W.; Huang, Y.J.; Leu, C.H.; Yeh, N.C.; Chu, C.Y.; Lin, C.C.; Shieh, G.S.; Chen, Y.L.; et al. Galectin-1 binds to influenza virus and ameliorates influenza virus pathogenesis. J. Virol. 2011, 85, 10010–10020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shim, J.A.; Park, S.; Lee, E.S.; Niki, T.; Hirashima, M.; Sohn, S. Galectin-9 ameliorates herpes simplex virus-induced inflammation through apoptosis. Immunobiology 2011, 217, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Garner, O.B.; Aguilar, H.C.; Fulcher, J.A.; Levroney, E.L.; Harrison, R.; Wright, L.; Robinson, L.R.; Aspericueta, V.; Panico, M.; Haslam, S.M.; et al. Endothelial galectin-1 binds to specific glycans on nipah virus fusion protein and inhibits maturation, mobility, and function to block syncytia formation. PLoS Pathog. 2010, 6, e1000993. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.Y.; Rabinovich, G.A.; Liu, F.T. Galectins: Structure, function and therapeutic potential. Expert Rev. Mol. Med. 2008, 10, e17. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Y.; Fermin, A.; Vardhana, S.; Weng, I.C.; Lo, K.F.; Chang, E.Y.; Maverakis, E.; Yang, R.Y.; Hsu, D.K.; Dustin, M.L.; et al. Galectin-3 negatively regulates TCR-mediated CD4+ T-cell activation at the immunological synapse. Proc. Natl. Acad. Sci. USA 2009, 106, 14496–14501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.H.; Lin, C.Y.; Chang, M.R.; Urbina, A.N.; Assavalapsakul, W.; Thitithanyanont, A.; Chen, Y.H.; Liu, F.T.; Wang, S.F. The role of galectins in virus infection—A systemic literature review. J. Microbiol. Immunol. Infect. 2019. [Google Scholar] [CrossRef] [PubMed]
- Fogel, S.; Guittaut, M.; Legrand, A.; Monsigny, M.; Hebert, E. The tat protein of HIV-1 induces galectin-3 expression. Glycobiology 1999, 9, 383–387. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.F.; Tsao, C.H.; Lin, Y.T.; Hsu, D.K.; Chiang, M.L.; Lo, C.H.; Chien, F.C.; Chen, P.; Arthur Chen, Y.M.; Chen, H.Y.; et al. Galectin-3 promotes HIV-1 budding via association with Alix and Gag p6. Glycobiology 2014, 24, 1022–1035. [Google Scholar] [CrossRef] [Green Version]
- Hsu, D.K.; Hammes, S.R.; Kuwabara, I.; Greene, W.C.; Liu, F.T. Human T lymphotropic virus-I infection of human T lymphocytes induces expression of the beta-galactoside-binding lectin, galectin-3. Am. J. Pathol. 1996, 148, 1661–1670. [Google Scholar]
- Fang, Z.; Xing, H.; Meng, Z.; Hong, K.; Liao, L.; He, X.; Shao, Y. Genetic characterization analysis of the tat exon-1 region of HIV type 1 CRF07_BC strains in China. AIDS Res. Hum. Retrovir. 2010, 26, 359–363. [Google Scholar] [CrossRef]
- Su, L.; Zhou, X.; Yuan, D.; Yang, H.; Wei, D.; Qin, G.; Liang, S. Prevalence and patterns of drug-resistance mutations among HIV-1 patients infected with CRF07_BC strains in Sichuan province, China. Virol. Sin. 2014, 29, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Hong, K.; Zhang, J.; Zhang, L.; Li, D.; Ren, L.; Liang, H.; Shao, Y. Construction and characterization of highly infectious full-length molecular clones of a HIV-1 CRF07_BC isolate from Xinjiang, China. PLoS ONE 2013, 8, e79177. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.H.; Meng, Z.F.; Xing, H.; Ruan, Y.H.; Li, X.P.; Xin, R.L.; Ma, P.F.; Peng, H.; Shao, Y. Analysis of HIV-1 CRF07_BC gag p6 sequences indicating novel deletions in the central region of p6. Arch. Virol. 2007, 152, 1553–1558. [Google Scholar] [CrossRef] [PubMed]
- Kanki, P.J.; Hamel, D.J.; Sankale, J.L.; Hsieh, C.; Thior, I.; Barin, F.; Woodcock, S.A.; Gueye-Ndiaye, A.; Zhang, E.; Montano, M.; et al. Human immunodeficiency virus type 1 subtypes differ in disease progression. J. Infect. Dis. 1999, 179, 68–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coetzer, M.; Cilliers, T.; Papathanasopoulos, M.; Ramjee, G.; Karim, S.A.; Williamson, C.; Morris, L. Longitudinal analysis of HIV type 1 subtype C envelope sequences from South Africa. AIDS Res. Hum. Retrovir. 2007, 23, 316–321. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, H.; Ren, X.; Wan, Z.; Hu, G.; Tang, S. HIV-1 CRF07_BC with a Seven Amino Acid Deletion in the gag p6 Region Dominates in HIV-1-Infected Men Who Have Sex with Men in China. AIDS Res. Hum. Retrovir. 2017, 33, 977–983. [Google Scholar] [CrossRef]
- Marlowe, N.; Flys, T.; Hackett, J., Jr.; Schumaker, M.; Jackson, J.B.; Eshleman, S.H.; Network, H.I.V.P.T.; Adult, A.C.T.G. Analysis of insertions and deletions in the gag p6 region of diverse HIV type 1 strains. AIDS Res. Hum. Retrovir. 2004, 20, 1119–11125. [Google Scholar] [CrossRef]
- Biesinger, T.; Kimata, J.T. HIV-1 Transmission, Replication Fitness and Disease Progression. Virology 2008, 2008, 49–63. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.W.; Li, W.Y.; Wang, W.H.; Lin, Y.T.; Chou, C.H.; Chen, M.; Huang, H.D.; Chen, Y.H.; Lu, P.L.; Wang, S.F.; et al. Characterization of the Drug Resistance Profiles of Patients Infected with CRF07_BC Using Phenotypic Assay and Ultra-Deep Pyrosequencing. PLoS ONE 2017, 12, e0170420. [Google Scholar] [CrossRef]
- Wu, H.; Zhang, H.J.; Zhang, X.M.; Xu, H.F.; Wang, M.; Huang, J.D.; Zheng, B.J. Identification of drug resistant mutations in HIV-1 CRF07_BC variants selected by nevirapine in vitro. PLoS ONE 2012, 7, e44333. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, R.; Prasad, A. Exosomes Derived from HIV-1 Infected DCs Mediate Viral trans-Infection via Fibronectin and Galectin-3. Sci. Rep. 2017, 7, 14787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrus, J.E.; von Schwedler, U.K.; Pornillos, O.W.; Morham, S.G.; Zavitz, K.H.; Wang, H.E.; Wettstein, D.A.; Stray, K.M.; Cote, M.; Rich, R.L.; et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 2001, 107, 55–65. [Google Scholar] [CrossRef] [Green Version]
- Carlton, J.G.; Agromayor, M.; Martin-Serrano, J. Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc. Natl. Acad. Sci. USA 2008, 105, 10541–10546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhai, Q.; Landesman, M.B.; Chung, H.Y.; Dierkers, A.; Jeffries, C.M.; Trewhella, J.; Hill, C.P.; Sundquist, W.I. Activation of the retroviral budding factor ALIX. J. Virol. 2011, 85, 9222–9226. [Google Scholar] [CrossRef] [Green Version]
- Usami, Y.; Popov, S.; Gottlinger, H.G. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J. Virol. 2007, 81, 6614–6622. [Google Scholar] [CrossRef] [Green Version]
- Sciacchitano, S.; Lavra, L.; Morgante, A.; Ulivieri, A.; Magi, F.; De Francesco, G.P.; Bellotti, C.; Salehi, L.B.; Ricci, A. Galectin-3: One Molecule for an Alphabet of Diseases, from A to Z. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Zhou, X.; Corvera, J.; Gallick, G.E.; Lin, S.H.; Kuang, J. ALG-2 activates the MVB sorting function of ALIX through relieving its intramolecular interaction. Cell Discov. 2015, 1, 15018. [Google Scholar] [CrossRef] [Green Version]
- Chan, Y.C.; Lin, H.Y.; Tu, Z.; Kuo, Y.H.; Hsu, S.D.; Lin, C.H. Dissecting the Structure-Activity Relationship of Galectin-Ligand Interactions. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef] [Green Version]
- Villard, R.; Hammache, D.; Delapierre, G.; Fotiadu, F.; Buono, G.; Fantini, J. Asymmetric synthesis of water-soluble analogues of galactosylceramide, an HIV-1 receptor: new tools to study virus-glycolipid interactions. Chembiochem 2002, 3, 517–525. [Google Scholar] [CrossRef]
- Lund, N.; Branch, D.R.; Mylvaganam, M.; Chark, D.; Ma, X.Z.; Sakac, D.; Binnington, B.; Fantini, J.; Puri, A.; Blumenthal, R.; et al. A novel soluble mimic of the glycolipid, globotriaosyl ceramide inhibits HIV infection. AIDS 2006, 20, 333–343. [Google Scholar] [CrossRef]
- Wei, M.; Guan, Q.; Liang, H.; Chen, J.; Chen, Z.; Hei, F.; Feng, Y.; Hong, K.; Huang, H.; Xing, H.; et al. Simple subtyping assay for human immunodeficiency virus type 1 subtypes B, C, CRF01-AE, CRF07-BC, and CRF08-BC. J. Clin. Microbiol. 2004, 42, 4261–4267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, Y.; Jia, W.; Kidoya, H.; Muramatsu, F.; Tsukada, Y.; Takakura, N. Galectin-3 Inhibits Cancer Metastasis by Negatively Regulating Integrin beta3 Expression. Am. J. Pathol. 2019, 189, 900–910. [Google Scholar] [CrossRef] [PubMed]



| HIV-1-Infected Population | |||
|---|---|---|---|
| Characteristics | Male (%) (n = 32) | Female (%) (n = 6) | Total (%) (n = 38) |
| Age (years) | |||
| 15–29 | 14(43.8) | 2(33.3) | 16(42.1) |
| 30–49 | 16(50) | 3(50) | 19(50) |
| ≥50 | 2(6.2) | 1(16.7) | 3(7.9) |
| Viral Load (copies/mL) | |||
| <10,000 | 2(6.2) | 1(16.7) | 3(7.8) |
| 10,000–100,000 | 19(59.4) | 3(50) | 22(57.9) |
| >100,000 | 11(34.4) | 2(33.3) | 13(34.2) |
| CD4 Count (cells/mm3) | |||
| <200 | 5(15.6) | 1(16.7) | 6(15.8) |
| 200–500 | 21(65.6) | 4(66.7) | 25(65.8) |
| >500 | 6(18.8) | 1(16.7) | 7(18.4) |
| Serum galectin-3 (mean ± SD) | 3.49 ± 0.51 | 3.24 ± 0.27 | 3.45 ± 0,49 |
| HIV-1 subtype | |||
| CRF01_AE | 2(6.3) | 3(50) | 5(13.2) |
| B | 7(21.9) | 1(16.7) | 8(22.2) |
| C | 0 | 0 | 0 |
| CRF07_BC | 23(71.9) | 2(33.3) | 25(69.4) |
| CRF08_BC | 0 | 0 | 0 |
| HIV-1 CRF07_BC Population | |||
|---|---|---|---|
| Virus Subtype | Male (%) (n = 23) | Female (%) (n = 2) | Total (%) (n = 25) |
| CRF07_BC | |||
| Gag-7d | 21(91.3) | 2(100) | 23(92) |
| Gag-11d | 2(8.7) | 0 | 2(8) |
| Gag-13d | 0 | 0 | 0 |
| Gag-wt | 0 | 0 | 0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.-H.; Yeh, C.-S.; Lin, C.-Y.; Yuan, R.-Y.; Urbina, A.N.; Lu, P.-L.; Chen, Y.-H.; Chen, Y.-M.A.; Liu, F.-T.; Wang, S.-F. Amino Acid Deletions in p6Gag Domain of HIV-1 CRF07_BC Ameliorate Galectin-3 Mediated Enhancement in Viral Budding. Int. J. Mol. Sci. 2020, 21, 2910. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21082910
Wang W-H, Yeh C-S, Lin C-Y, Yuan R-Y, Urbina AN, Lu P-L, Chen Y-H, Chen Y-MA, Liu F-T, Wang S-F. Amino Acid Deletions in p6Gag Domain of HIV-1 CRF07_BC Ameliorate Galectin-3 Mediated Enhancement in Viral Budding. International Journal of Molecular Sciences. 2020; 21(8):2910. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21082910
Chicago/Turabian StyleWang, Wen-Hung, Chun-Sheng Yeh, Chih-Yen Lin, Ruei-Yu Yuan, Aspiro Nayim Urbina, Po-Liang Lu, Yen-Hsu Chen, Yi-Ming Arthur Chen, Fu-Tong Liu, and Sheng-Fan Wang. 2020. "Amino Acid Deletions in p6Gag Domain of HIV-1 CRF07_BC Ameliorate Galectin-3 Mediated Enhancement in Viral Budding" International Journal of Molecular Sciences 21, no. 8: 2910. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21082910





