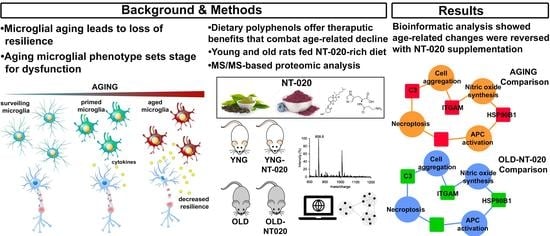
Polyphenol Supplementation Reverses Age-Related Changes in Microglial Signaling Cascades
Abstract
:1. Introduction
2. Results/Discussion
2.1. NT-020 Supplementation Alters Microglial Protein Expression in Aging Rats
2.2. Gene Ontology Analysis Highlights Global Functional Changes in Aging Tat Microglial Proteome with NT-020 Diet
2.3. Pathway Analysis Identifies Canonical Pathways Driving Age-Related Changes in Microglial Proteome Reversed with NT-020 Diet
2.4. NT-020 Diet Reverses Age-Related Changes in Predicted Rat Microglia Function
3. Conclusions
4. Materials and Methods
4.1. Animal Protocol and NT-020 Supplementation
4.2. Microglial Extraction and Cell Culture
4.3. Sample Lysis, In-Gel Digest, and Label-Free Quantification
4.4. Simple Western
4.5. Gene Ontology Analysis with GOplot in R
4.6. Proteomic Comparisons in IPA
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hou, Y.; Dan, X.; Babbar, M.; Wei, Y.; Hasselbalch, S.G.; Croteau, D.L.; Bohr, V.A. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 2019, 15, 565–581. [Google Scholar] [CrossRef] [PubMed]
- Norden, D.M.; Godbout, J.P. Review: Microglia of the aged brain: Primed to be activated and resistant to regulation. Neuropathol. Appl. Neurobiol. 2012, 39, 19–34. [Google Scholar] [CrossRef]
- Colton, C.A. Heterogeneity of Microglial Activation in the Innate Immune Response in the Brain. J. Neuroimmune Pharmacol. 2009, 4, 399–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Barres, B.A. Microglia and macrophages in brain homeostasis and disease. Nat. Rev. Immunol. 2018, 18, 225–242. [Google Scholar] [CrossRef]
- Wake, H.; Moorhouse, A.J.; Jinno, S.; Kohsaka, S.; Nabekura, J. Resting Microglia Directly Monitor the Functional State of Synapses In Vivo and Determine the Fate of Ischemic Terminals. J. Neurosci. 2009, 29, 3974–3980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parkhurst, C.N.; Gan, W.-B. Microglia dynamics and function in the CNS. Curr. Opin. Neurobiol. 2010, 20, 595–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durafourt, B.A.; Moore, C.S.; Zammit, D.A.; Johnson, T.A.; Zaguia, F.; Guiot, M.-C.; Bar-Or, A.; Antel, J.P. Comparison of polarization properties of human adult microglia and blood-derived macrophages. Glia 2012, 60, 717–727. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.A.; Das, A.; Ray, S.K.; Banik, N.L. Role of pro-inflammatory cytokines released from microglia in neurodegenerative diseases. Brain Res. Bull. 2012, 87, 10–20. [Google Scholar] [CrossRef]
- Fu, R.; Shen, Q.; Xu, P.; Luo, J.J.; Tang, Y. Phagocytosis of Microglia in the Central Nervous System Diseases. Mol. Neurobiol. 2014, 49, 1422–1434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Bernhardi, R.; Eugenín-von Bernhardi, L.; Eugenín, J. Microglial cell dysregulation in brain aging and neurodegeneration. Front. Aging Neurosci. 2015, 7, 124. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.-G.; Ding, J.-Q.; Chen, S.-D. Microglia in the aging brain: Relevance to neurodegeneration. Mol. Neurodegener. 2010, 5, 12. [Google Scholar] [CrossRef] [Green Version]
- Mandrekar-Colucci, S. Microglia and Inflammation in Alzheimers Disease. CNS Neurol. Disord. Drug Targets 2010, 9, 156–167. [Google Scholar] [CrossRef]
- Boillée, S.; Yamanaka, K.; Lobsiger, C.S.; Copeland, N.G.; Jenkins, N.A.; Kassiotis, G.; Kollias, G.; Cleveland, D.W. Onset and Progression in Inherited ALS Determined by Motor Neurons and Microglia. Science 2006, 312, 1389–1392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Wang, T.; Pei, Z.; Miller, D.S.; Wu, X.; Block, M.L.; Wilson, B.; Zhang, W.; Zhou, Y.; Hong, J.-S.; et al. Aggregated α-synuclein activates microglia: A process leading to disease progression in Parkinson’s disease. FASEB J. 2005, 19, 533–542. [Google Scholar] [CrossRef] [PubMed]
- Flowers, A.; Bell-Temin, H.; Jalloh, A.; Stevens, S.M.; Bickford, P.C. Proteomic analysis of aged microglia: Shifts in transcription, bioenergetics, and nutrient response. J. Neuroinflamm. 2017, 14, 96. [Google Scholar] [CrossRef] [PubMed]
- Frank, M.G.; Barrientos, R.M.; Biedenkapp, J.C.; Rudy, J.W.; Watkins, L.R.; Maier, S.F. mRNA up-regulation of MHC II and pivotal pro-inflammatory genes in normal brain aging. Neurobiol. Aging 2006, 27, 717–722. [Google Scholar] [CrossRef] [PubMed]
- Stichel, C.; Luebbert, H. Inflammatory processes in the aging mouse brain: Participation of dendritic cells and T-cells. Neurobiol. Aging 2007, 28, 1507–1521. [Google Scholar] [CrossRef] [PubMed]
- Sierra, A.; Gottfried-Blackmore, A.C.; McEwen, B.S.; Bulloch, K. Microglia derived from aging mice exhibit an altered inflammatory profile. Glia 2007, 55, 412–424. [Google Scholar] [CrossRef] [PubMed]
- Kitazawa, M.; Oddo, S.; Yamasaki, T.R.; Green, K.N.; LaFerla, F.M. Lipopolysaccharide-Induced Inflammation Exacerbates Tau Pathology by a Cyclin-Dependent Kinase 5-Mediated Pathway in a Transgenic Model of Alzheimer’s Disease. J. Neurosci. 2005, 25, 8843–8853. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.C.; Rizer, J.; Selenica, M.-L.B.; Reid, P.; Kraft, C.; Johnson, A.; Blair, L.; Gordon, M.N.; Dickey, C.A.; Morgan, D. LPS- induced inflammation exacerbates phospho-tau pathology in rTg4510 mice. J. Neuroinflamm. 2010, 7, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiu, H.-F.; Venkatakrishnan, K.; Wang, C.-K. The role of nutraceuticals as a complementary therapy against various neurodegenerative diseases: A mini-review. J. Tradit. Complement. Med. 2020, 10, 434–439. [Google Scholar] [CrossRef]
- Liu, H.-W.; Chan, Y.-C.; Wang, M.-F.; Wei, C.-C.; Chang, S.-J. Dietary (−)-Epigallocatechin-3-gallate Supplementation Counteracts Aging-Associated Skeletal Muscle Insulin Resistance and Fatty Liver in Senescence-Accelerated Mouse. J. Agric. Food Chem. 2015, 63, 8407–8417. [Google Scholar] [CrossRef] [PubMed]
- Sarubbo, F.; Ramis, M.R.; Kienzer, C.; Aparicio, S.; Esteban, S.; Miralles, A.; Moranta, D. Chronic Silymarin, Quercetin and Naringenin Treatments Increase Monoamines Synthesis and Hippocampal Sirt1 Levels Improving Cognition in Aged Rats. J. Neuroimmune Pharmacol. 2018, 13, 24–38. [Google Scholar] [CrossRef] [PubMed]
- Calvello, R.; Cianciulli, A.; Nicolardi, G.; De Nuccio, F.; Giannotti, L.; Salvatore, R.; Porro, C.; Trotta, T.; Panaro, M.A.; Lofrumento, D.D. Vitamin D Treatment Attenuates Neuroinflammation and Dopaminergic Neurodegeneration in an Animal Model of Parkinson’s Disease, Shifting M1 to M2 Microglia Responses. J. Neuroimmune Pharmacol. 2016, 12, 327–339. [Google Scholar] [CrossRef]
- Mukherjee, B.S.; Mk, P.; Poddar, M.K. Carnosine restores aging-induced elevation of corticosterone status and brain regional amyloid-beta in relation to down regulation of locomotor activity. J. Syst. Integr. Neurosci. 2019, 5, 1–11. [Google Scholar] [CrossRef]
- Bickford, P.C.; Tan, J.; Shytle, R.D.; Sanberg, C.D.; El-Badri, N.; Sanberg, P.R. Nutraceuticals Synergistically Promote Proliferation of Human Stem Cells. Stem Cells Dev. 2006, 15, 118–123. [Google Scholar] [CrossRef]
- Yasuhara, T.; Hara, K.; Maki, M.; Masuda, T.; Sanberg, C.D.; Sanberg, P.R.; Bickford, P.C.; Borlongan, C.V. Dietary Supplementation Exerts Neuroprotective Effects in Ischemic Stroke Model. Rejuvenation Res. 2008, 11, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Acosta, S.; Jernberg, J.; Sanberg, C.D.; Sanberg, P.R.; Small, B.J.; Gemma, C.; Bickford, P.C. NT-020, a Natural Therapeutic Approach to Optimize Spatial Memory Performance and Increase Neural Progenitor Cell Proliferation and Decrease Inflammation in the Aged Rat. Rejuvenation Res. 2010, 13, 581–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flowers, A.; Lee, J.-Y.; Acosta, S.; Hudson, C.; Small, B.; Sanberg, C.D.; Bickford, P.C.; Grimmig, B. NT-020 treatment reduces inflammation and augments Nrf-2 and Wnt signaling in aged rats. J. Neuroinflamm. 2015, 12, 174. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Lu, B.; Sheng, L.; Zhu, Z.; Sun, H.; Zhou, Y.; Yang, Y.; Xue, D.; Chen, W.; Tian, X.; et al. Hexokinase 2-dependent hyperglycolysis driving microglial activation contributes to ischemic brain injury. J. Neurochem. 2018, 144, 186–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al Mamun, A.; Chauhan, A.; Qi, S.; Ngwa, C.; Xu, Y.; Sharmeen, R.; Hazen, A.L.; Li, J.; Aronowski, J.A.; McCullough, L.D.; et al. Microglial IRF5-IRF4 regulatory axis regulates neuroinflammation after cerebral ischemia and impacts stroke outcomes. Proc. Natl. Acad. Sci. USA 2020, 117, 1742–1752. [Google Scholar] [CrossRef] [PubMed]
- D’Ambrosi, N.; Rossi, S.; Gerbino, V.; Cozzolino, M.; D’Ambrosi, N. Rac1 at the crossroad of actin dynamics and neuroinflammation in Amyotrophic Lateral Sclerosis. Front. Cell. Neurosci. 2014, 8. [Google Scholar] [CrossRef] [Green Version]
- Kawai, K.; Tsuno, N.H.; Kitayama, J.; Okaji, Y.; Yazawa, K.; Asakage, M.; Hori, N.; Watanabe, T.; Takahashi, K.; Nagawa, H. Epigallocatechin gallate attenuates adhesion and migration of CD8+ T cells by binding to CD11b. J. Allergy Clin. Immunol. 2004, 113, 1211–1217. [Google Scholar] [CrossRef] [PubMed]
- Kawai, K.; Tsuno, N.H.; Kitayama, J.; Sunami, E.; Takahashi, K.; Nagawa, H. Catechin inhibits adhesion and migration of peripheral blood B cells by blocking CD11b. Immunopharmacol. Immunotoxicol. 2010, 33, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Wang, W.; Wang, D.; Ling, W. Quercetin protects against atherosclerosis by inhibiting dendritic cell activation. Mol. Nutr. Food Res. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.W.; Selhorst, A.; Lampe, S.G.; Liu, Y.; Yang, Y.; Lovett-Racke, A.E. Neuron-Specific Vitamin D Signaling Attenuates Microglia Activation and CNS Autoimmunity. Front. Neurol. 2020, 11, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caruso, G.; Fresta, C.G.; Musso, N.; Giambirtone, M.; Grasso, M.; Spampinato, S.F.; Merlo, S.; Drago, F.; Lazzarino, G.; Sortino, M.A.; et al. Carnosine Prevents Aβ-Induced Oxidative Stress and Inflammation in Microglial Cells: A Key Role of TGF-β1. Cells 2019, 8, 64. [Google Scholar] [CrossRef] [Green Version]
- Reimand, J.; Kull, M.; Peterson, H.; Hansen, J.; Vilo, J. g: Profiler—A web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007, 35, W193–W200. [Google Scholar] [CrossRef]
- Scheper, W.; Hoozemans, J.J. The unfolded protein response in neurodegenerative diseases: A neuropathological perspective. Acta Neuropathol. 2015, 130, 315–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.H.; Won, S.M.; Suh, J.; Son, S.J.; Moon, G.J.; Park, U.-J.; Gwag, B.J. Induction of the unfolded protein response and cell death pathway in Alzheimer’s disease, but not in aged Tg2576 mice. Exp. Mol. Med. 2010, 42, 386–394. [Google Scholar] [CrossRef] [Green Version]
- Hoozemans, J.J.M.; Van Haastert, E.S.; Nijholt, D.A.T.; Rozemuller, A.J.M.; Scheper, W. Activation of the Unfolded Protein Response Is an Early Event in Alzheimer’s and Parkinson’s Disease. Neurodegener. Dis. 2012, 10, 212–215. [Google Scholar] [CrossRef] [PubMed]
- Martínez, G.; Duran-Aniotz, C.; Cabral-Miranda, F.; Vivar, J.P.; Hetz, C. Endoplasmic reticulum proteostasis impairment in aging. Aging Cell 2017, 16, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Van Beek, A.A.; Bossche, J.V.D.; Mastroberardino, P.G.; de Winther, M.P.; Leenen, P.J. Metabolic Alterations in Aging Macrophages: Ingredients for Inflammaging? Trends Immunol. 2019, 40, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Martucciello, S.; Masullo, M.; Cerulli, A.; Piacente, S. Natural Products Targeting ER Stress, and the Functional Link to Mitochondria. Int. J. Mol. Sci. 2020, 21, 1905. [Google Scholar] [CrossRef] [Green Version]
- Ettcheto, M.; Cano, A.; Manzine, P.R.; Busquets, O.; Verdaguer, E.; Castro-Torres, R.D.; García, M.L.; Beas-Zarate, C.; Olloquequi, J.; Auladell, C.; et al. Epigallocatechin-3-Gallate (EGCG) Improves Cognitive Deficits Aggravated by an Obesogenic Diet Through Modulation of Unfolded Protein Response in APPswe/PS1dE9 Mice. Mol. Neurobiol. 2019, 57, 1814–1827. [Google Scholar] [CrossRef]
- Ehrlich, L.C.; Hu, S.; Sheng, W.S.; Sutton, R.L.; Rockswold, G.L.; Peterson, P.K.; Chao, C.C. Cytokine regulation of human microglial cell IL-8 production. J. Immunol. 1998, 160, 1944–1948. [Google Scholar]
- Ryu, J.K.; Cho, T.; Choi, H.B.; Jantaratnotai, N.; McLarnon, J.G. Pharmacological antagonism of interleukin-8 receptor CXCR2 inhibits inflammatory reactivity and is neuroprotective in an animal model of Alzheimer’s disease. J. Neuroinflamm. 2015, 12, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Di Rosa, M.; Malaguarnera, G.; De Gregorio, C.; Palumbo, M.; Nunnari, G.; Malaguarnera, L. Immuno-modulatory effects of vitamin D3 in human monocyte and macrophages. Cell. Immunol. 2012, 280, 36–43. [Google Scholar] [CrossRef]
- Chu, C.; Liu, L.; Wang, Y.; Yang, R.; Hu, C.; Rung, S.; Man, Y.; Qu, Y. Evaluation of epigallocatechin-3-gallate (EGCG)-modified scaffold determines macrophage recruitment. Mater. Sci. Eng. C 2019, 100, 505–513. [Google Scholar] [CrossRef]
- Hu, Y.; Mai, W.; Chen, L.; Cao, K.; Zhang, B.; Zhang, Z.; Liu, Y.; Lou, H.; Duan, S.; Gao, Z. mTOR-mediated metabolic reprogramming shapes distinct microglia functions in response to lipopolysaccharide and ATP. Glia 2019, 68, 1031–1045. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, C.; Yao, Y.; Chen, L.; Liu, G.; Zhang, R.; Liu, Q.; Shi, F.; Hao, J. mTORC1 pathway disruption ameliorates brain inflammation following stroke via a shift in microglia phenotype from M1 type to M2 type. FASEB J. 2016, 30, 3388–3399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russo, C.D.; Lisi, L.; Tringali, G.; Navarra, P. Involvement of mTOR kinase in cytokine-dependent microglial activation and cell proliferation. Biochem. Pharmacol. 2009, 78, 1242–1251. [Google Scholar] [CrossRef] [Green Version]
- Giampieri, F.; Afrin, S.; Forbes-Hernandez, T.Y.; Gasparrini, M.; Cianciosi, D.; Rodríguez, P.R.; Varela-López, A.; Quiles, J.L.; Battino, M. Autophagy in Human Health and Disease: Novel Therapeutic Opportunities. Antioxid. Redox Signal. 2019, 30, 577–634. [Google Scholar] [CrossRef]
- Gu, H.-F.; Nie, Y.-X.; Tong, Q.-Z.; Tang, Y.-L.; Zeng, Y.; Jing, K.-Q.; Zheng, X.-L.; Liao, D.-F. Epigallocatechin-3-Gallate Attenuates Impairment of Learning and Memory in Chronic Unpredictable Mild Stress-Treated Rats by Restoring Hippocampal Autophagic Flux. PLoS ONE 2014, 9, e112683. [Google Scholar] [CrossRef] [PubMed]
- Regitz, C.; Dußling, L.M.; Wenzel, U. Amyloid-beta (Aβ1-42)-induced paralysis inCaenorhabditis elegansis inhibited by the polyphenol quercetin through activation of protein degradation pathways. Mol. Nutr. Food Res. 2014, 58, 1931–1940. [Google Scholar] [CrossRef] [PubMed]
- Frank, M.G.; Barrientos, R.M.; Watkins, L.R.; Maier, S.F. Aging sensitizes rapidly isolated hippocampal microglia to LPS ex vivo. J. Neuroimmunol. 2010, 226, 181–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, K.; Zhao, Y.-F. Aging modulates microglia phenotypes in neuroinflammation of MPTP-PD mice. Exp. Gerontol. 2018, 111, 86–93. [Google Scholar] [CrossRef]
- Damani, M.R.; Zhao, L.; Fontainhas, A.M.; Amaral, J.; Fariss, R.N.; Wong, W.T. Age-related alterations in the dynamic behavior of microglia. Aging Cell 2010, 10, 263–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Z.; Zhou, T.; Sun, X.; Zheng, Y.; Cheng, B.; Li, M.; Liu, X.; He, C. Necroptosis in microglia contributes to neuroinflammation and retinal degeneration through TLR4 activation. Cell Death Differ. 2018, 25, 180–189. [Google Scholar] [CrossRef] [Green Version]
- Byun, E.-B.; Choi, H.-G.; Sung, N.-Y. Green tea polyphenol epigallocatechin-3-gallate inhibits TLR4 signaling through the 67-kDa laminin receptor on lipopolysaccharide-stimulated dendritic cells. Biochem. Biophys. Res. Commun. 2012, 426, 480–485. [Google Scholar] [CrossRef]
- Lin, S.-K.; Chang, H.-H.; Chen, Y.-J.; Wang, C.-C.; Galson, D.L.; Hong, C.-Y.; Kok, S.-H.; Chen, A.Y.-J. Epigallocatechin-3-gallate diminishes CCL2 expression in human osteoblastic cells via up-regulation of phosphatidylinositol 3-Kinase/Akt/Raf-1 interaction: A potential therapeutic benefit for arthritis. Arthritis Rheum. 2008, 58, 3145–3156. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Tang, H.-B.; Shan, L.-Q.; Liu, S.-C.; Huang, D.-G.; Chen, X.; Chen, Z.; Yang, M.; Yin, X.-H.; Yang, H.; et al. Quercetin prevents necroptosis of oligodendrocytes by inhibiting macrophages/microglia polarization to M1 phenotype after spinal cord injury in rats. J. Neuroinflamm. 2019, 16, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhao, Y.; Zhu, X.; Guo, Y.; Yang, Y.; Jiang, Y.; Liu, B. Active vitamin D regulates macrophage M1/M2 phenotypes via the STAT-1-TREM-1 pathway in diabetic nephropathy. J. Cell. Physiol. 2019, 234, 6917–6926. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Rodríguez, C.; Martín-Sanz, E.; Cuadrado, E.; Granizo, J.J.; Sanz-Fernández, R. Protective effect of polyphenols on presbycusis via oxidative/nitrosative stress suppression in rats. Exp. Gerontol. 2016, 83, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Bensalem, J.; Dudonné, S.; Gaudout, D.; Servant, L.; Calon, F.; Desjardins, Y.; Layé, S.; Lafenetre, P.; Pallet, V. Polyphenol-rich extract from grape and blueberry attenuates cognitive decline and improves neuronal function in aged mice. J. Nutr. Sci. 2018, 7, e19. [Google Scholar] [CrossRef] [Green Version]
- Ammar, A.; Trabelsi, K.; Boukhris, O.; Bouaziz, B.; Müller, P.; Glenn, J.M.; Bott, N.T.; Müller, N.; Chtourou, H.; Driss, T.; et al. Effects of Polyphenol-Rich Interventions on Cognition and Brain Health in Healthy Young and Middle-Aged Adults: Systematic Review and Meta-Analysis. J. Clin. Med. 2020, 9, 1598. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.-C.; Clark, D.J.; Schnaubelt, M.; Teo, G.C.; Leprevost, F.D.V.; Bocik, W.; Boja, E.S.; Hiltke, T.; Nesvizhskii, A.I.; Zhang, H. Deep Proteomics Using Two Dimensional Data Independent Acquisition Mass Spectrometry. Anal. Chem. 2020, 92, 4217–4225. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jalloh, A.; Flowers, A.; Hudson, C.; Chaput, D.; Guergues, J.; Stevens, S.M., Jr.; Bickford, P.C. Polyphenol Supplementation Reverses Age-Related Changes in Microglial Signaling Cascades. Int. J. Mol. Sci. 2021, 22, 6373. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22126373
Jalloh A, Flowers A, Hudson C, Chaput D, Guergues J, Stevens SM Jr., Bickford PC. Polyphenol Supplementation Reverses Age-Related Changes in Microglial Signaling Cascades. International Journal of Molecular Sciences. 2021; 22(12):6373. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22126373
Chicago/Turabian StyleJalloh, Ahmad, Antwoine Flowers, Charles Hudson, Dale Chaput, Jennifer Guergues, Stanley M. Stevens, Jr., and Paula C. Bickford. 2021. "Polyphenol Supplementation Reverses Age-Related Changes in Microglial Signaling Cascades" International Journal of Molecular Sciences 22, no. 12: 6373. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22126373