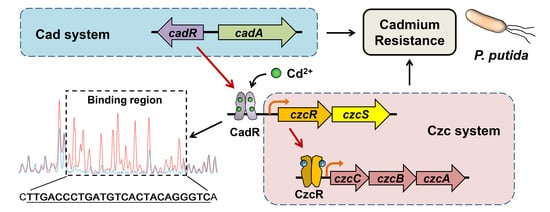
The Connection between Czc and Cad Systems Involved in Cadmium Resistance in Pseudomonas putida
Abstract
:1. Introduction
2. Results and Discussion
2.1. Involvement of Two-Component System CzcRS3 in Heavy Metal Resistance
2.2. CzcRS3 Controls the Expression of Two CzcCBA Efflux Pumps
2.3. CzcR3 Directly Binds to the Promoters of czcCBA1 and czcCBA2
2.4. Identification of the Transcription Pattern of czcRS3
2.5. Cd2+ Induces czcRS3 Expression through CadR
2.6. CadR Binds to czcRS3 Promoter Directly
3. Materials and Methods
3.1. Bacterial Strains and Growth Conditions
3.2. Construction of Strains and Plasmids
3.3. Test for Susceptibility to Heavy Metals
3.4. Measurement of β-Galactosidase Activity
3.5. Identification of Transcription Start Site
3.6. Purification of His-Tagged CzcR3 and CadR
3.7. Generation of Fluorescent Probes of Promoters
3.8. Electrophoretic Mobility Shift Assay (EMSA)
3.9. DNase I Footprinting Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, F.; Chen, X.; Liu, Y.; Niu, Z.; Tang, H.; Mao, S.; Li, N.; Chen, G.; Xiang, H. Serum cardiovascular-related metabolites disturbance exposed to different heavy metal exposure scenarios. J. Hazard. Mater. 2021, 415, 125590. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, R.; Li, Y.; Ding, X.; Jiang, Y.; Feng, J.; Zhu, L. Trophic transfer of heavy metals in the marine food web based on tissue residuals. Sci. Total. Environ. 2021, 772, 145064. [Google Scholar] [CrossRef]
- Sun, Z.; Xie, X.; Wang, P.; Hu, Y.; Cheng, H. Heavy metal pollution caused by small-scale metal ore mining activities: A case study from a polymetallic mine in South China. Sci. Total. Environ. 2018, 639, 217–227. [Google Scholar] [CrossRef]
- Huttunen-Saarivirta, E.; Korpiniemi, H.; Kuokkala, V.T.; Paajanen, H. Corrosion of cadmium plating by runway de-icing chemicals: Study of surface phenomena and comparison of corrosion tests. Surf. Coat. Tech. 2013, 232, 101–115. [Google Scholar] [CrossRef]
- Xiao, J.; Huang, J.; Wang, M.; Huang, M.; Wang, Y. The fate and long-term toxic effects of NiO nanoparticles at environmental concentration in constructed wetland: Enzyme activity, microbial property, metabolic pathway and functional genes. J. Hazard. Mater. 2021, 413, 125295. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Adebayo, A.; Jia, J.L.; Xing, Y.; Deng, S.Q.; Guo, L.M.; Liang, Y.T.; Zhang, D.Y. Impacts of heavy metals and soil properties at a Nigerian e-waste site on soil microbial community. J. Hazard. Mater. 2019, 362, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Nies, D.H. Microbial heavy-metal resistance. Appl. Microbiol. Biot. 1999, 51, 730–750. [Google Scholar] [CrossRef]
- Smith, R.L.; Maguire, M.E. Distribution of the cora Mg(2+) transport-system in Gram-negative bacteria. J. Bacteriol. 1995, 177, 1638–1640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grass, G.; Wong, M.D.; Rosen, B.P.; Smith, R.L.; Rensing, C. ZupT is a Zn(II) uptake system in Escherichia coli. J. Bacteriol. 2002, 184, 864–866. [Google Scholar] [CrossRef] [Green Version]
- Que, Q.; Helmann, J.D. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol. Microbiol. 2000, 35, 1454–1468. [Google Scholar] [CrossRef]
- Ammendola, S.; Cerasi, M.; Battistoni, A. Deregulation of transition metals homeostasis is a key feature of cadmium toxicity in Salmonella. Biometals 2014, 27, 703–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Cobbina, S.J.; Mao, G.; Xu, H.; Zhang, Z.; Yang, L. A review of toxicity and mechanisms of individual and mixtures of heavy metals in the environment. Environ. Sci. Pollut. Res. Int. 2016, 23, 8244–8259. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.J.; Kim, Y.S.; Kumar, V. Heavy metal toxicity: An update of chelating therapeutic strategies. J. Trace Elem. Med. Biol. 2019, 54, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.M.; Hu, W.Y.; Wang, H.F.; Liu, P.; Wang, X.K.; Huang, B.A. Spatial distribution, ecological risk and sources of heavy metals in soils from a typical economic development area, Southeastern China. Sci. Total. Environ. 2021, 780, 146557. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.K.; Huang, B.; Hu, W.Y.; Wang, W.X.; Muhammad, I.; Lu, Q.Q.; Jing, G.H.; Zhang, Z. Ecological-health risks assessment and source identification of heavy metals in typical greenhouse vegetable production systems in Northwest China. Environ. Sci. Pollut. Res. 2021, 28, 42583–42595. [Google Scholar] [CrossRef]
- Comte, S.; Guibaud, G.; Baudu, M. Biosorption properties of extracellular polymeric substances (EPS) towards Cd, Cu and Pb for different pH values. J. Hazard. Mater. 2008, 151, 185–193. [Google Scholar] [CrossRef]
- Blindauer, C.A. Bacterial metallothioneins: Past, present, and questions for the future. J. Biol. Inorg. Chem. 2011, 16, 1011–1024. [Google Scholar] [CrossRef]
- Gallardo-Benavente, C.; Carrion, O.; Todd, J.D.; Pieretti, J.C.; Seabra, A.B.; Duran, N.; Rubilar, O.; Perez-Donoso, J.M.; Quiroz, A. Biosynthesis of CdS quantum dots mediated by volatile sulfur compounds released by antarctic Pseudomonas fragi. Front. Microbiol. 2019, 10, 1866. [Google Scholar] [CrossRef] [Green Version]
- Shi, Z.J.; Zhang, Z.F.; Yuan, M.; Wang, S.; Yang, M.; Yao, Q.; Ba, W.N.; Zhao, J.; Xie, B. Characterization of a high cadmium accumulating soil bacterium, Cupriavidus sp. WS2. Chemosphere 2020, 247, 125834. [Google Scholar] [CrossRef]
- Nies, D.H. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 2003, 27, 313–339. [Google Scholar] [CrossRef]
- Nanda, M.; Kumar, V.; Sharma, D.K. Multimetal tolerance mechanisms in bacteria: The resistance strategies acquired by bacteria that can be exploited to ‘clean-up’ heavy metal contaminants from water. Aquat. Toxicol. 2019, 212, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.C.; Zheng, X.; Zhang, D.N.; Iqbal, W.; Liu, C.K.; Yang, B.; Zhao, X.; Lu, X.Y.; Mao, Y.P. Microbial characterization of heavy metal resistant bacterial strains isolated from an electroplating wastewater treatment plant. Ecotox. Environ. Safe 2019, 181, 472–480. [Google Scholar] [CrossRef]
- Xia, X.; Wu, S.J.; Zhou, Z.J.; Wang, G.J. Microbial Cd(II) and Cr(VI) resistance mechanisms and application in bioremediation. J. Hazard. Mater. 2021, 401, 123685. [Google Scholar] [CrossRef] [PubMed]
- Rensing, C.; Pribyl, T.; Nies, D.H. New functions for the three subunits of the CzcCBA cation-proton antiporter. J. Bacteriol. 1997, 179, 6871–6879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nies, D.H. The cobalt, zinc, and cadmium efflux system CzcABC from Alcaligenes eutrophus functions as a cation-proton antiporter in Escherichia coli. J. Bacteriol. 1995, 177, 2707–2712. [Google Scholar] [CrossRef] [Green Version]
- Dieppois, G.; Ducret, V.; Caille, O.; Perron, K. The transcriptional regulator CzcR modulates antibiotic resistance and quorum sensing in Pseudomonas aeruginosa. PLoS ONE 2012, 7, e38148. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Chen, W.; Huang, S.; He, Y.; Liu, X.; Hu, Q.; Wei, T.; Sang, H.; Gan, J.; Chen, H. Structural basis of Zn(II) induced metal detoxification and antibiotic resistance by histidine kinase CzcS in Pseudomonas aeruginosa. PLoS Pathog. 2017, 13, e1006533. [Google Scholar] [CrossRef] [Green Version]
- Anton, A.; Grosse, C.; Reissmann, J.; Pribyl, T.; Nies, D.H. CzcD is a heavy metal ion transporter involved in regulation of heavy metal resistance in Ralstonia sp. strain CH34. J. Bacteriol. 1999, 181, 6876–6881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnawi, H.; Masri, N.; Hussain, N.; Al-Lawati, B.; Mayasari, E.; Gulbicka, A.; Jervis, A.J.; Huang, M.H.; Cavet, J.S.; Linton, D. RNA-based thermoregulation of a Campylobacter jejuni zinc resistance determinant. PLoS Pathog. 2020, 16, e1009008. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Hu, Q.; Yang, J.; Huang, S.; Wei, T.; Chen, W.; He, Y.; Wang, D.; Liu, Z.; Wang, K.; et al. Selective cadmium regulation mediated by a cooperative binding mechanism in CadR. Proc. Natl. Acad. Sci. USA 2019, 116, 20398–20403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.C.; Gardarin, A.; Martel, A.; Mintz, E.; Guillain, F.; Catty, P. The cadmium transport sites of CadA, the Cd2+-ATPase from Listeria monocytogenes. J. Biol. Chem. 2006, 281, 29533–29541. [Google Scholar] [CrossRef] [Green Version]
- Endo, G.; Silver, S. CadC, the transcriptional regulatory protein of the cadmium resistance system of Staphylococcus aureus plasmid pI258. J. Bacteriol. 1995, 177, 4437–4441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Legatzki, A.; Grass, G.; Anton, A.; Rensing, C.; Nies, D.H. Interplay of the Czc system and two P-type ATPases in conferring metal resistance to Ralstonia metallidurans. J. Bacteriol. 2003, 185, 4354–4361. [Google Scholar] [CrossRef] [Green Version]
- Leedjarv, A.; Ivask, A.; Virta, M. Interplay of different transporters in the mediation of divalent heavy metal resistance in Pseudomonas putida KT2440. J. Bacteriol. 2008, 190, 2680–2689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin-Pascual, M.; Batianis, C.; Bruinsma, L.; Asin-Garcia, E.; Garcia-Morales, L.; Weusthuis, R.A.; van Kranenburg, R.; Martins Dos Santos, V.A.P. A navigation guide of synthetic biology tools for Pseudomonas putida. Biotechnol. Adv. 2021, 49, 107732. [Google Scholar] [CrossRef]
- Zhao, Y.X.; Che, Y.; Zhang, F.; Wang, J.C.; Gao, W.X.; Zhang, T.; Yang, C. Development of an efficient pathway construction strategy for rapid evolution of the biodegradation capacity of Pseudomonas putida KT2440 and its application in bioremediation. Sci. Total. Environ. 2021, 761, 143239. [Google Scholar] [CrossRef]
- Fernandez-Lopez, C.; Posada-Baquero, R.; Garcia, J.L.; Castilla-Alcantara, J.C.; Cantos, M.; Ortega-Calvo, J.J. Root-mediated bacterial accessibility and cometabolism of pyrene in soil. Sci. Total. Environ. 2021, 760, 143408. [Google Scholar] [CrossRef] [PubMed]
- Kohlstedt, M.; Starck, S.; Barton, N.; Stolzenberger, J.; Selzer, M.; Mehlmann, K.; Schneider, R.; Pleissner, D.; Rinkel, J.; Dickschat, J.S.; et al. From lignin to nylon: Cascaded chemical and biochemical conversion using metabolically engineered Pseudomonas putida. Metab. Eng. 2018, 47, 279–293. [Google Scholar] [CrossRef]
- Nikel, P.I.; de Lorenzo, V. Pseudomonas putida as a functional chassis for industrial biocatalysis: From native biochemistry to trans-metabolism. Metab. Eng. 2018, 50, 142–155. [Google Scholar] [CrossRef]
- Hu, N.; Zhao, B. Key genes involved in heavy-metal resistance in Pseudomonas putida CD2. FEMS Microbiol. Lett. 2007, 267, 17–22. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.L.; Chen, X.; Huang, Q.Y.; Chen, W.L. The role of CzcRS two-component systems in the heavy metal resistance of Pseudomonas putida X4. Int. J. Mol. Sci. 2015, 16, 17005–17017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagdasarian, M.; Lurz, R.; Ruckert, B.; Franklin, F.C.H.; Bagdasarian, M.M.; Frey, J.; Timmis, K.N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 1981, 16, 237–247. [Google Scholar] [CrossRef]
- Winsor, G.L.; Griffiths, E.J.; Lo, R.; Dhillon, B.K.; Shay, J.A.; Brinkman, F.S.L. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016, 44, D646–D653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canovas, D.; Cases, I.; de Lorenzo, V. Heavy metal tolerance and metal homeostasis in Pseudomonas putida as revealed by complete genome analysis. Environ. Microbiol. 2003, 5, 1242–1256. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, R.; Sahoo, B.K.; Saini, D.K. Cross-talk and specificity in two-component signal transduction pathways. Future Microbiol. 2016, 11, 685–697. [Google Scholar] [CrossRef]
- Brocker, M.; Mack, C.; Bott, M. Target genes, consensus binding site, and role of phosphorylation for the response regulator MtrA of Corynebacterium glutamicum. J. Bacteriol. 2011, 193, 1237–1249. [Google Scholar] [CrossRef] [Green Version]
- Jeon, Y.; Lee, Y.S.; Han, J.S.; Kim, J.B.; Hwang, D.S. Multimerization of phosphorylated and non-phosphorylated ArcA is necessary for the response regulator function of the Arc two-component signal transduction system. J. Biol. Chem. 2001, 276, 40873–40879. [Google Scholar] [CrossRef] [Green Version]
- Brown, D.R.; Barton, G.; Pan, Z.S.; Buck, M.; Wigneshweraraj, S. Nitrogen stress response and stringent response are coupled in Escherichia coli. Nat. Commun. 2014, 5, 4115. [Google Scholar] [CrossRef] [Green Version]
- Garber, M.E.; Rajeev, L.; Kazakov, A.E.; Trinh, J.; Masuno, D.; Thompson, M.G.; Kaplan, N.; Luk, J.; Novichkov, P.S.; Mukhopadhyay, A. Multiple signaling systems target a core set of transition metal homeostasis genes using similar binding motifs. Mol. Microbiol. 2018, 107, 704–717. [Google Scholar] [CrossRef] [Green Version]
- Typas, A.; Becker, G.; Hengge, R. The molecular basis of selective promoter activation by the σS subunit of RNA polymerase. Mol. Microbiol. 2007, 63, 1296–1306. [Google Scholar] [CrossRef]
- Zhang, X.X.; Rainey, P.B. Dual involvement of CbrAB and NtrBC in the regulation of histidine utilization in Pseudomonas fluorescens SBW25. Genetics 2008, 178, 185–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.W.; Glickmann, E.; Cooksey, D.A. Chromosomal locus for cadmium resistance in Pseudomonas putida consisting of a cadmium-transporting ATPase and a MerR family response regulator. Appl. Environ. Microb. 2001, 67, 1437–1444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, W.; Liu, Y.; Giometti, C.S.; Tollaksen, S.L.; Khare, T.; Wu, L.; Klingeman, D.M.; Fields, M.W.; Zhou, J. Knock-out of SO1377 gene, which encodes the member of a conserved hypothetical bacterial protein family COG2268, results in alteration of iron metabolism, increased spontaneous mutation and hydrogen peroxide sensitivity in Shewanella oneidensis MR-1. BMC Genom. 2006, 7, 76. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Yan, H.; Xiao, Y.; Nie, H.; Huang, Q.; Chen, W. The exopolysaccharide gene cluster pea is transcriptionally controlled by RpoS and repressed by AmrZ in Pseudomonas putida KT2440. Microbiol. Res. 2019, 218, 1–11. [Google Scholar] [CrossRef]
- Liu, H.Z.; Xiao, Y.J.; Nie, H.L.; Huang, Q.Y.; Chen, W.L. Influence of (p)ppGpp on biofilm regulation in Pseudomonas putida KT2440. Microbiol. Res. 2017, 204, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Nie, H.L.; Xiao, Y.J.; Liu, H.Z.; He, J.Z.; Chen, W.L.; Huang, Q.Y. FleN and FleQ play a synergistic role in regulating lapA and bcs operons in Pseudomonas putida KT2440. Env. Microbiol. Rep. 2017, 9, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Miyake, Y.; Fujiwara, S.; Usui, T.; Suginaka, H. Simple method for measuring the antibiotic concentration required to kill adherent bacteria. Chemotherapy 1992, 38, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.H. Experiments in Molecular Genetics; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1972. [Google Scholar]
- Liu, F.; Zheng, K.; Chen, H.C.; Liu, Z.F. Capping-RACE: A simple, accurate, and sensitive 5’ RACE method for use in prokaryotes. Nucleic Acids Res. 2018, 46, e129. [Google Scholar] [CrossRef] [Green Version]







| Metal Ion (mM) | Cd2+ | Co2+ | Cr3+ | Cu2+ | Mn2+ | Ni2+ | Pb2+ | Zn2+ |
|---|---|---|---|---|---|---|---|---|
| wild-type | 1 | 1 | 4 | 8 | 16 | 8 | 8 | 8 |
| ΔczcRS3 | 0.25 | 1 | 4 | 8 | 16 | 8 | 8 | 8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Zhang, Y.; Wang, Y.; Xie, X.; Shi, Q. The Connection between Czc and Cad Systems Involved in Cadmium Resistance in Pseudomonas putida. Int. J. Mol. Sci. 2021, 22, 9697. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22189697
Liu H, Zhang Y, Wang Y, Xie X, Shi Q. The Connection between Czc and Cad Systems Involved in Cadmium Resistance in Pseudomonas putida. International Journal of Molecular Sciences. 2021; 22(18):9697. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22189697
Chicago/Turabian StyleLiu, Huizhong, Yu Zhang, Yingsi Wang, Xiaobao Xie, and Qingshan Shi. 2021. "The Connection between Czc and Cad Systems Involved in Cadmium Resistance in Pseudomonas putida" International Journal of Molecular Sciences 22, no. 18: 9697. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22189697