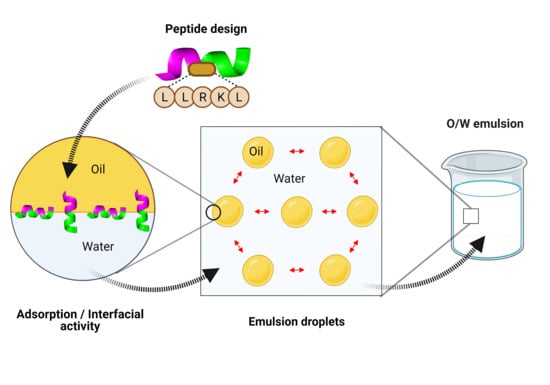
Emerging Emulsifiers: Conceptual Basis for the Identification and Rational Design of Peptides with Surface Activity
Abstract
:1. Introduction
2. Emulsion Formation
3. Molecular Characteristics of Emulsifying Peptides
| Peptide Sequence | Evaluation Method | Value | References |
|---|---|---|---|
| Ac-MKQLADS LHQLARQ VSRLEHA-CONH2 | Surface tension at 25 °C, pH 7.4 and a peptide concentration of 5 µM (mN/m) | 52 | [59] |
| Ac-MKQLADS LHQLAHK VSHLEHA-CONH2 | 53.1 | [59] | |
| Ac-MKQLADS LMQLARQ VSRLESA-CONH2 | 51.5 | [59] | |
| Ac-LMQLARQ-MKQLADS-LMQLARQ-VSRLESA-CONH2 | Interfacial tension (mN/m) in the system octane-water. The concentration of the peptide was 4.5 µM and the interfacial tension without peptide was 51 mN/m. | 13.5 | [60] |
| YF | γCMC (mN/m) at 25 °C | 48.5 | [46] |
| VTV | 52.3 | [46] | |
| FEFRFEFR | 52.5 | [61] | |
| FEFKFEFK | 47 | [61] | |
| AEAKAEAKAEAKAEAK | 57 | [62] | |
| LEELLEELLEELLEEL | Surface tension (mN/m) at pH 7, a peptide concentration of 0.001% (w/v) and 25 °C | 56.8 | [15] |
| Surface tension (mN/m) at pH 5.5, a peptide concentration of 0.001% (w/v) and 25 °C | 40.51 | [15] | |
| ELELELELELELELEL | Surface tension (mN/m) at pH 7, a peptide concentration of 0.001% (w/v) and 25 °C | 54.3 | [15] |
| Surface tension (mN/m) at pH 5.5, a peptide concentration of 0.001% (w/v) and 25 °C | 49.51 | [15] | |
| LELLEEELLEEELLEL | Surface tension (mN/m) at pH 7, a peptide concentration of 0.001% (w/v) and 25 °C | 65.5 | [15] |
| Surface tension (mN/m) at pH 5.5, a peptide concentration of 0.001% (w/v) and 25 °C | 55.01 | [15] | |
| RELEELNVPGEIVESLSSSEESITR | Surface tension at pH 7 (mN/m). The concentration of the peptide was 0.05% (w/v) | 56.1 | [55] |
| Surface tension at pH 3 (mN/m). The concentration of the peptide was 0.05% (w/v) | 51.8 | ||
| YQEPVLGPVRGPFPIIV | Surface tension at pH 7 (mN/m). The concentration of the peptide was 0.05% (w/v) | 54.4 | [55] |
| Surface tension at pH 3 (mN/m). The concentration of the peptide was 0.05% (w/v) | 50.1 | ||
| LSFNPTQLEEQCHI | Presence at the interface of the hexadecane−water system at 20 °C | Present | [54] |
| YSLAMAASDISLLDAQSAPLRVYVEELKPTPEGDLEILLQKW | Present | [54] | |
| SLAMAASDISLL | Present | [54] | |
| VYVEELKPTPEGDLEIL | Present | [54] | |
| VYVEELKPTPEGDLEILLQK | Present | [54,63] | |
| WENGECAQK | Presence at the oil−protein aqueous solution interface | Present | [63] |
| IIAEK | Present | [63] | |
| IDALNENK | Present | [63] | |
| VLVLDTDYKK | Present | [63] | |
| ALK | Present | [63] | |
| ALPMHIR | Present | [63] | |
| LIVTQTMK | Present | [63] | |
| GKNHDTGVSPVFA | Interfacial tension (mN/m) for the system: 25% dodecane 75% crude oil (w/v)/aqueous solution at pH 7 and 1 M of NaCl. The concentration of the peptide was 550 ppm in the oil phase and the clean interface interfacial tension value was 45 mN/m | 30 | [64] |
| DPKDGSVVVL | 42.7 | [64] | |
| TGNTCDNVKQR | 32 | [64] | |
| THENQLGAGAFG | 37.5 | [64,65] | |
| QRAALIDCLAPDRRV | 39 | [64] | |
| QRAALIDCLA | 38 | [64] | |
| ILEFLEGQLQEVDN | Interfacial tension (mN/m) with Medium Chain Triglycerides (MCT) oil. A peptide concentration 0.2%wt. was used in the aqueous phase with an adjusted pH of 7 with a buffer solution. The interfacial tension of the clean interface was 26 mN/m | 22.41 | [18] |
| KYDGKYLMQVLQE | 18.86 | [18] | |
| KYLMQVLQEKL | 17.87 | [18] | |
| KKPVSKDSPETYEEALKRFAKLLSDRKKL | 16.93 | [18] | |
| EALKRFAKLLSD | 16.83 | [18] | |
| AKDIVPFYFEHGPHIFN | 16.63 | [18] | |
| KYLMQVLQEKLGE | 16.48 | [18] | |
| IPATILEFLEGQLQEVDNN | 15.4 | [18] | |
| DSPETYEEALKRFAKLLSD | 14.28 | [18] | |
| NRPFAAAKDIVPFYFEHGPHIFN | 14.13 | [18] | |
| DDNFCAKVGVVIQ | 24.72 | [18] | |
| LNIQFNI | 22.04 | [18] | |
| GKELDPRLSYRI | 21.12 | [18] | |
| LGGDVYLGKSPNSDAPCP | 19.33 | [18] | |
| VHQNGKRRLALVKDNPLDVSFK | 18.11 | [18] | |
| FIPLSTNIFEDQLLNIQFNIPT | 14.89 | [18] | |
| CPFSSDDQFCLKVGV | 13.97 | [18] | |
| IGSSSHFGPHIFEGELLNIQFDIS | 13.62 | [18] | |
| ELDSRLSYRIISTFWGALGGDVYLGKSPN | 12.12 | [18] | |
| ELDSRLSYRIISTFWGALGGDVYL | 12.09 | [18] | |
| LNIQFNIPTPKLC | 12.07 | [18] | |
| TPNENNRPFAAAKDIV | 23.11 | [18] | |
| FAKLLSDRKKLRANK | 21.81 | [18] | |
| VGVVIQNGKRR | 21.34 | [18] | |
| NPNSSYRIISI | 20.78 | [18] | |
| CRDDNFCAKVGVVI | 20.14 | [18] | |
| LLTAMITTPNENNRP | 19.67 | [18] | |
| RDDNFCAKVGVVI | 18.15 | [18] | |
| DNFCAKVGVVIQNGKRR | 18.16 | [18] | |
| FCLKVGVIHQNGKRRLALVK | 17.64 | [18] | |
| DTNGKELNPNSSYRIISIGRGALGGDVYL | 17.41 | [18] | |
| FCLKVGVVHQNGKRRLALVKDNP | 17.26 | [18] | |
| HQNGKRRLALV | 15.26 | [18] | |
| SSDDQFCLKVGVV | 15.07 | [18] | |
| FDVIGGTSTGGLLTAMITTPNENNRP | 13.73 | [18] | |
| KDNPETYEEALKRFAKLLS | 13.74 | [18] | |
| GIIPATILEFLEGQLQEVDNN | 13.22 | [18] | |
| GIKGIIPAIILEFLEGQLQEVDNNKDAR | 10.84 | [18] | |
| IRPIPFIPRGGKT-NH2 | γCMC (mN/m) at 25 °C | 72.01 | [66] |
| FIGALLRPALKLLA-NH2 | 46.9 | [66] | |
| GLKEVAHSAKKFAKGFISGLTGS | 72.01 | [66] | |
| LKGASKLIPHLLPSRQQ | 72.01 | [66] | |
| FIGALLGPLLNLLK-NH2 | 38.3 | [66] | |
| IRPVPFFPPVHAKKVFPLH | 72.01 | [66] |
4. Impact of Emulsion Conditions on the Peptide Emulsifying Behavior
5. Methodologies for Screening and Designing Emulsifying Peptides
5.1. Identification of Emulsifying Peptides from Hydrolyzed Extracts of Proteins
5.2. Design of Emulsifying Peptides from Proteins through Computational Tools
5.3. Design De Novo of Emulsifying Peptides
5.4. Identification of Emulsifying Peptides by Mimetization
5.5. Design of Emulsifying Peptides by the Crosslinking of Peptides at the Interface
6. Challenges and Opportunities
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hack, B.; Egger, H.; Uhlemann, J.; Henriet, M.; Wirth, W.; Vermeer, A.W.P.; Duff, D. Advanced agrochemical formulations through encapsulation strategies? Chem. Ing. Tech. 2012, 84, 223–234. [Google Scholar] [CrossRef]
- Cussler, E.L.; Moggridge, G.D. Chemical Product Design, 2nd ed.; Cambridge University Press: Cambridge, UK, 2011. [Google Scholar]
- Mattei, M.; Kontogeorgis, G.M.; Gani, R. A comprehensive framework for surfactant selection and design for emulsion based chemical product design. Fluid Phase Equilib. 2014, 362, 288–299. [Google Scholar] [CrossRef]
- Rosen, M.J. Surfactants and Interfacial Phenomena, 3rd ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2004; ISBN 0471478180. [Google Scholar]
- Khan, B.A.; Akhtar, N.; Khan, H.M.S.; Waseem, K.; Mahmood, T.; Rasul, A.; Iqbal, M.; Khan, H. Basics of pharmaceutical emulsions: A review. Afr. J. Pharm. Pharmacol. 2011, 5, 2715–2725. [Google Scholar] [CrossRef]
- Rehfeld, S.J. The effects of initial surfactant concentration and emulsification time upon the particle size and distribution of benzene-in-water emulsions. J. Colloid Interface Sci. 1967, 24, 358–365. [Google Scholar] [CrossRef]
- NG, K.M.; Gani, R.; Dam-Johansen, K. Chemical Product Design: Towards a Perspective through Case Studies. In Chapter 1: Chemical Product Design—A Brief Overview; NG, K.M., Gani, R., Dam-Johansen, K., Eds.; Elsevier Science: Amsterdam, The Netherlands, 2007. [Google Scholar]
- Usaid, A. Emulsion and it’s Applications in Food Processing-A Review. Int. J. Eng. Res. Appl. 2014, 4, 241–248. [Google Scholar]
- Akbari, S.; Abdurahman, N.H.; Yunus, R.M.; Fayaz, F.; Alara, O.R. Biosurfactants—a new frontier for social and environmental safety: A mini review. Biotechnol. Res. Innov. 2018, 2, 81–90. [Google Scholar] [CrossRef]
- Rufino, R.D.; de Luna, J.M.; de Campos Takaki, G.M.; Sarubbo, L.A. Characterization and properties of the biosurfactant produced by Candida lipolytica UCP 0988. Electron. J. Biotechnol. 2014, 17, 34–38. [Google Scholar] [CrossRef] [Green Version]
- Tao, W.; Lin, J.; Wang, W.; Huang, H.; Li, S. Designer bioemulsifiers based on combinations of different polysaccharides with the novel emulsifying esterase AXE from Bacillus subtilis CICC 20034. Microb. Cell Fact. 2019, 18, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Hayes, D.G. Biobased Surfactants Overview and Industrial State-of-the-Art. In Biobased Surfactants and Detergents; Hayes, D.G., Kitamoto, D., Solaiman, D., Ashby, R., Eds.; AOCS Press: Urbana, IL, USA, 2009; pp. 3–25. ISBN 978-1-893997-67-7. [Google Scholar]
- Smith, B.V.; Ierapepritou, M.G. Integrative chemical product design strategies: Reflecting industry trends and challenges. Comput. Chem. Eng. 2010, 34, 857–865. [Google Scholar] [CrossRef]
- Cardona Jaramillo, J.E.C.; Achenie, L.E.; Álvarez, O.A.; Carrillo Bautista, M.P.; González Barrios, A.F. The multiscale approach to the design of bio-based emulsions. Curr. Opin. Chem. Eng. 2020, 27, 65–71. [Google Scholar] [CrossRef]
- Saito, M.; Ogasawara, M.; Chikuni, K.; Shimizu, M. Synthesis of a Peptide Emulsifier with an Amphiphilic Structure. Biosci. Biotechnol. Biochem. 1995, 59, 388–392. [Google Scholar] [CrossRef]
- McClements, D.J.; Bai, L.; Chung, C. Recent Advances in the Utilization of Natural Emulsifiers to Form and Stabilize Emulsions. Annu. Rev. Food Sci. Technol. 2017, 8, 205–236. [Google Scholar] [CrossRef]
- Uzoigwe, C.; Burgess, J.G.; Ennis, C.J.; Rahman, P.K.S.M. Bioemulsifiers are not biosurfactants and require different screening approaches. Front. Microbiol. 2015, 6, 245. [Google Scholar] [CrossRef] [Green Version]
- García-Moreno, P.J.; Gregersen, S.; Nedamani, E.R.; Olsen, T.H.; Marcatili, P.; Overgaard, M.T.; Andersen, M.L.; Hansen, E.B.; Jacobsen, C. Identification of emulsifier potato peptides by bioinformatics: Application to omega-3 delivery emulsions and release from potato industry side streams. Sci. Rep. 2020, 10, 690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poon, S.; Clarke, A.E.; Schultz, C.J. Structure-function analysis of the emulsifying and interfacial properties of apomyoglobin and derived peptides. J. Colloid Interface Sci. 1999, 213, 193–203. [Google Scholar] [CrossRef]
- De Faria, A.F.; Teodoro-Martinez, D.S.; De Oliveira Barbosa, G.N.; Gontijo Vaz, B.; Serrano Silva, Í.; Garcia, J.S.; Tótola, M.R.; Eberlin, M.N.; Grossman, M.; Alves, O.L.; et al. Production and structural characterization of surfactin (C 14/Leu7) produced by Bacillus subtilis isolate LSFM-05 grown on raw glycerol from the biodiesel industry. Process Biochem. 2011, 46, 1951–1957. [Google Scholar] [CrossRef] [Green Version]
- McClements, D.J.; Gumus, C.E. Natural emulsifiers—Biosurfactants, phospholipids, biopolymers, and colloidal particles: Molecular and physicochemical basis of functional performance. Adv. Colloid Interface Sci. 2016, 234, 3–26. [Google Scholar] [CrossRef] [Green Version]
- Dave, N.; Joshi, T. A Concise Review on Surfactants and Its Significance. Int. J. Appl. Chem. 2017, 13, 663–672. [Google Scholar]
- Costa, C.; Medronho, B.; Filipe, A.; Mira, I.; Lindman, B.; Edlund, H.; Norgren, M. Emulsion formation and stabilization by biomolecules: The leading role of cellulose. Polymers 2019, 11, 1570. [Google Scholar] [CrossRef] [Green Version]
- Holmberg, K.; Jönsson, B.; Kronberg, B.; Lindman, B. Surfactant and Polymers in Aqueous Solution, 2nd ed.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2003; ISBN 0471498831. [Google Scholar]
- Farn, R.J. Chemistry and Technology of Surfactants, 1st ed.; Blackwell Publishing Ltd: Oxford, UK, 2007; ISBN 1405126965. [Google Scholar]
- Tadros, T. Encyclopedia of Colloid and Interface Science; Springer Reference: Berlin, Germany, 2013. [Google Scholar]
- Kong, X.; Zhou, H.; Qian, H. Enzymatic preparation and functional properties of wheat gluten hydrolysates. Food Chem. 2007, 101, 615–620. [Google Scholar] [CrossRef]
- Malcolm, A.; Dexter, A.; Middelberg, A. Peptide surfactants (Pepfactants) for switchable foams and emulsions. Asia-Pac. J. Chem. Eng. 2007, 2, 362–367. [Google Scholar] [CrossRef]
- McClements, D.J. Critical review of techniques and methodologies for characterization of emulsion stability. Crit. Rev. Food Sci. Nutr. 2007, 47, 611–649. [Google Scholar] [CrossRef]
- Huibers, P.D.T.; Lobanov, V.S.; Katritzky, A.R.; Shah, D.O.; Karelson, M. Prediction of critical micelle concentration using a quantitative structure-property relationship approach. 1. Nonionic surfactants. Langmuir 1996, 12, 1462–1470. [Google Scholar] [CrossRef]
- Márquez, N.; Bravo, B.; Ysambertt, F.; Chávez, G.; Subero, N.; Salager, J.L. Analysis of polyethoxylated surfactants in microemulsion-oil-water systems: III. Fractionation and partitioning of polyethoxylated alcohol surfactants. Anal. Chim. Acta 2003, 477, 293–303. [Google Scholar] [CrossRef]
- Nakamura, A.; Muramatsu, M. Complex formation between N-dodecyl-β-alanine and sodium alkylsulfate as reflected in coadsorption from mixed solution. J. Colloid Interface Sci. 1977, 62, 165–171. [Google Scholar] [CrossRef]
- Belton, J.; Twidle, H. The Surface Tensions of Amino-Acid Solutions. Trans. Faraday Soc. 1940, 36, 1198–1208. [Google Scholar] [CrossRef]
- Rodríguez, D.M.; Romero, C.M. Surface Tension of Glycine, Alanine, Aminobutyric Acid, Norvaline, and Norleucine in Water and in Aqueous Solutions of Strong Electrolytes at Temperatures from (293.15 to 313.15) K. J. Chem. Eng. Data 2017, 62, 3687–3696. [Google Scholar] [CrossRef]
- Raza, M.A.; Hallett, P.D.; Liu, X.; He, M.; Afzal, W. Surface Tension of Aqueous Solutions of Small-Chain Amino and Organic Acids. J. Chem. Eng. Data 2019, 64, 5049–5056. [Google Scholar] [CrossRef]
- Yiase, S.G. Amino acids as potential emulsifiers in stabilizing oil/water emulsions. Int. J. Innov. Sci. Res. 2015, 15, 409–414. [Google Scholar]
- Black, C.D.; Popovich, N.G. A study of intravenous emulsion compatibility: Effects of dextrose, amino acids, and selected electrolytes. Drug Intell. Clin. Pharm. 1981, 15, 184–193. [Google Scholar] [CrossRef]
- Imura, T.; Nakayama, M.; Taira, T.; Sakai, H.; Abe, M.; Kitamoto, D. Interfacial and emulsifying properties of soybean peptides with different degrees of hydrolysis. J. Oleo Sci. 2015, 64, 183–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sewald, N.; Jakubke, H.-D. Peptides: Chemistry and Biology, 1st ed.; Wiley-VCH: Weinheim, Germany, 2002. [Google Scholar]
- Fauchère, J.-L.; Pliska, V. Hydrophobic parameters II of amino acid side-chains from the partitioning of N-acetyl-amino acid amides. Eur. J. Med. Chem. 1983, 18, 369–375. [Google Scholar]
- Li, J.; Wang, J.; Zhao, Y.; Zhou, P.; Carter, J.; Li, Z.; Waigh, T.A.; Lu, J.R.; Xu, H. Surfactant-like peptides: From molecular design to controllable self-assembly with applications. Coord. Chem. Rev. 2020, 421, 213418–213432. [Google Scholar] [CrossRef]
- Edwards-Gayle, C.J.C.; Hamley, I.W. Self-assembly of bioactive peptides, peptide conjugates, and peptide mimetic materials. Org. Biomol. Chem. 2017, 15, 5867–5876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, Y.; Tu, B.; Yu, L.; Zheng, Y.; Lin, Y.; Luo, W.; Yang, Y.; Fang, Q.; Wang, C. Peptide conformation and oligomerization characteristics of surface-mediated assemblies revealed by molecular dynamics simulations and scanning tunneling microscopy. RSC Adv. 2019, 9, 41345–41350. [Google Scholar] [CrossRef] [Green Version]
- Xue, Y.; He, L.; Middelberg, A.P.J.; Mark, A.E.; Poger, D. Determining the structure of interfacial peptide films: Comparing neutron reflectometry and molecular dynamics simulations. Langmuir 2014, 30, 10080–10089. [Google Scholar] [CrossRef]
- Dexter, A.F.; Middelberg, A.P.J. Peptides as functional surfactants. Ind. Eng. Chem. Res. 2008, 47, 6391–6398. [Google Scholar] [CrossRef]
- James, J.; Mandal, A.B. Micelle formation of Tyr-Phe dipeptide and Val-Tyr-Val tripeptide in aqueous solution and their influence on the aggregation of SDS and PEO-PPO-PEO copolymer micelles. Colloids Surf. B Biointerfaces 2011, 84, 172–180. [Google Scholar] [CrossRef]
- Scholberg, H.M.; Guenthner, R.A.; Coon, R.I. Surface chemistry of fluorocarbons and their derivatives. J. Phys. Chem. 1953, 57, 923–925. [Google Scholar] [CrossRef]
- Middelberg, A.P.J.; Dexter, A.F. Peptide Networks. International Patent WO2006089364A1, 31 August 2016. [Google Scholar]
- Saiani, A.; Mohammed, A.; Frielinghaus, H.; Collins, R.; Hodson, N.; Kielty, C.M.; Sherratt, M.J.; Miller, A.F. Self-assembly and gelation properties of α-helix versus β-sheet forming peptides. Soft Matter 2009, 5, 193–202. [Google Scholar] [CrossRef]
- Hamley, I.W. Self-assembly of amphiphilic peptides. Soft Matter 2011, 7, 4122–4138. [Google Scholar] [CrossRef] [Green Version]
- Bai, S.; Pappas, C.; Debnath, S.; Frederix, P.W.J.M.; Leckie, J.; Fleming, S.; Ulijn, R.V. Stable emulsions formed by self-assembly of interfacial networks of dipeptide derivatives. ACS Nano 2014, 8, 7005–7013. [Google Scholar] [CrossRef] [PubMed]
- Scott, G.G.; McKnight, P.J.; Tuttle, T.; Ulijn, R.V. Tripeptide Emulsifiers. Adv. Mater. 2016, 28, 1381–1386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Girardet, J.M.; Debomy, L.; Courthaudon, J.L.; Miclo, L.; Humbert, G.; Gaillard, J.L. Viscoelastic properties of oil-water interfaces covered by bovine β-casein tryptic peptides. J. Dairy Sci. 2000, 83, 2410–2421. [Google Scholar] [CrossRef]
- Rahali, V.; Chobert, J.M.; Haertlé, T.; Guéguen, J. Emulsification of chemical and enzymatic hydrolysates of β-lactoglobulin: Characterization of the peptides adsorbed at the interface. Nahr. Food 2000, 44, 89–95. [Google Scholar] [CrossRef]
- Lee, S.W.; Shimizu, M.; Kaminogawa, S.; Yamauchi, K. Emulsifying properties of peptides obtained from the hydrolyzates of β-casein. Agric. Biol. Chem. 1987, 51, 161–166. [Google Scholar] [CrossRef] [Green Version]
- Dickinson, E. Properties of Emulsions Stabilized with Milk Proteins: Overview of Some Recent Developments. J. Dairy Sci. 1997, 80, 2607–2619. [Google Scholar] [CrossRef]
- Davis, J.P.; Doucet, D.; Foegeding, E.A. Foaming and interfacial properties of hydrolyzed β-lactoglobulin. J. Colloid Interface Sci. 2005, 288, 412–422. [Google Scholar] [CrossRef] [PubMed]
- Kilara, A.; Panyam, D. Peptides from Milk Proteins and Their Properties. Crit. Rev. Food Sci. Nutr. 2003, 43, 607–633. [Google Scholar] [CrossRef] [PubMed]
- Dexter, A.F.; Middelberg, A.P.J. Switchable peptide surfactants with designed metal binding capacity. J. Phys. Chem. C 2007, 111, 10484–10492. [Google Scholar] [CrossRef]
- Middelberg, A.P.J.; Radke, C.J.; Blanch, H.W. Peptide interfacial adsorption is kinetically limited by the thermodynamic stability of self association. Proc. Natl. Acad. Sci. USA 2000, 97, 5054–5059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, H.L.; Curtis, R. Inter-relation of surface tension and optical turbidity in self-assembled peptide amphiphiles. Biointerface Res. Appl. Chem. 2017, 7, 1913–1920. [Google Scholar]
- Hong, Y.; Lau, L.S.; Legge, R.L.; Chen, P. Critical self-assembly concentration of an ionic-complementary peptide EAK16-I. J. Adhes. 2004, 80, 913–931. [Google Scholar] [CrossRef]
- Persaud, D.R.; Dalgleish, D.G.; Nadeau, L.; Gauthier, S. Isolation and purification of serum and interfacial peptides of a trypsinolyzed β-lactoglobulin oil-in-water emulsion. J. Chromatogr. B Biomed. Sci. Appl. 2000, 744, 389–397. [Google Scholar] [CrossRef]
- Fernández-Niño, M.; Rojas, L.; Cifuentes, J.; Torres, R.; Ordonez, A.; Cruz, J.C.; Vargas, E.F.; Pradilla, D.; Solano, O.Á.; Barrios, A.G. Insights into the behavior of six rationally designed peptides based on Escherichia coli’s OmpA at the water-dodecane interface. PLoS ONE 2019, 14, e0223670. [Google Scholar] [CrossRef]
- Aguilera-Segura, S.M.; Núñez Vélez, V.; Achenie, L.; Álvarez Solano, O.; Torres, R.; González Barrios, A.F. Peptides design based on transmembrane Escherichia coli’s OmpA protein through molecular dynamics simulations in water–dodecane interfaces. J. Mol. Graph. Model. 2016, 68, 216–223. [Google Scholar] [CrossRef]
- Shigeri, Y.; Yasuda, A.; Hagihara, Y.; Nishi, K.; Watanabe, K.; Imura, T.; Inagaki, H.; Haramoto, Y.; Ito, Y.; Asashima, M. Identification of novel peptides from amphibian (Xenopus tropicalis) skin by direct tissue MALDI-MS analysis. FEBS J. 2015, 282, 102–113. [Google Scholar] [CrossRef] [Green Version]
- Acquah, C.; Di Stefano, E.; Udenigwe, C.C. Role of hydrophobicity in food peptide functionality and bioactivity. J. Food Bioact. 2018, 4, 88–98. [Google Scholar] [CrossRef] [Green Version]
- Perez-Riverol, Y.; Audain, E.; Millan, A.; Ramos, Y.; Sanchez, A.; Vizcaíno, J.A.; Wang, R.; Müller, M.; Machado, Y.J.; Betancourt, L.H.; et al. Isoelectric point optimization using peptide descriptors and support vector machines. J. Proteom. 2012, 75, 2269–2274. [Google Scholar] [CrossRef]
- Turgeon, S.L.; Gauthier, S.F.; Paquin, P. Emulsifying Property of Whey Peptide Fractions as a Function of pH and ionic Strength. J. Food Sci. 1992, 57, 601–604. [Google Scholar] [CrossRef]
- Lam, R.S.H.; Nickerson, M.T. Food proteins: A review on their emulsifying properties using a structure-function approach. Food Chem. 2013, 141, 975–984. [Google Scholar] [CrossRef]
- Alashi, A.M.; Blanchard, C.L.; Mailer, R.J.; Agboola, S.O.; Mawson, J.A.; Aluko, R.E. Influence of enzymatic hydrolysis, pH and storage temperature on the emulsifying properties of canola protein isolate and hydrolysates. Int. J. Food Sci. Technol. 2018, 53, 2316–2324. [Google Scholar] [CrossRef]
- Lee, S.J.; Choi, S.J.; Li, Y.; Decker, E.A.; McClements, D.J. Protein-stabilized nanoemulsions and emulsions: Comparison of physicochemical stability, lipid oxidation, and lipase digestibility. J. Agric. Food Chem. 2011, 59, 415–427. [Google Scholar] [CrossRef] [PubMed]
- Keowmaneechai, E.; McClements, D.J. Effect of CaCl2 and KCl on physiochemical properties of model nutritional beverages based on whey protein stabilized oil-in-water emulsions. J. Food Sci. 2002, 67, 665–671. [Google Scholar] [CrossRef]
- Adnoju, R. Whey protein peptides as dual-functional ingredients in food nanoemulsions. Ph.D. Thesis, University of Ghana, Legon, Ghana, 2014. [Google Scholar]
- Martínez-Maqueda, D.; Miralles, B.; Cruz-Huerta, E.; Recio, I. Casein hydrolysate and derived peptides stimulate mucin secretion and gene expression in human intestinal cells. Int. Dairy J. 2013, 32, 13–19. [Google Scholar] [CrossRef]
- Claustre, J.; Toumi, F.; Trompette, A.; Jourdan, G.; Guignard, H.; Chayvialle, J.A.; Plaisancié, P. Effects of peptides derived from dietary proteins on mucus secretion in rat jejunum. Am. J. Physiol. Gastrointest. Liver Physiol. 2002, 283, 521–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cudennec, B.; Ravallec-Plé, R.; Courois, E.; Fouchereau-Peron, M. Peptides from fish and crustacean by-products hydrolysates stimulate cholecystokinin release in STC-1 cells. Food Chem. 2008, 111, 970–975. [Google Scholar] [CrossRef]
- El Hatmi, H.; Jrad, Z.; Khorchani, T.; Jardin, J.; Poirson, C.; Perrin, C.; Cakir-Kiefer, C.; Girardet, J.M. Identification of bioactive peptides derived from caseins, glycosylation-dependent cell adhesion molecule-1 (GlyCAM-1), and peptidoglycan recognition protein-1 (PGRP-1) in fermented camel milk. Int. Dairy J. 2016, 56, 159–168. [Google Scholar] [CrossRef]
- Balti, R.; Bougatef, A.; Sila, A.; Guillochon, D.; Dhulster, P.; Nedjar-Arroume, N. Nine novel angiotensin I-converting enzyme (ACE) inhibitory peptides from cuttlefish (Sepia officinalis) muscle protein hydrolysates and antihypertensive effect of the potent active peptide in spontaneously hypertensive rats. Food Chem. 2015, 170, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Lafarga, T.; Aluko, R.E.; Rai, D.K.; O’Connor, P.; Hayes, M. Identification of bioactive peptides from a papain hydrolysate of bovine serum albumin and assessment of an antihypertensive effect in spontaneously hypertensive rats. Food Res. Int. 2016, 81, 91–99. [Google Scholar] [CrossRef] [Green Version]
- Lin, P.; Wong, J.H.; Ng, T.B. A defensin with highly potent antipathogenic activities from the seeds of purple pole bean. Biosci. Rep. 2010, 30, 101–109. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Dong, S.; Xu, J.; Zeng, M.; Song, H.; Zhao, Y. Production of cysteine-rich antimicrobial peptide by digestion of oyster (Crassostrea gigas) with alcalase and bromelin. Food Control 2008, 19, 231–235. [Google Scholar] [CrossRef]
- Popineau, Y.; Huchet, B.; Larré, C.; Bérot, S. Foaming and emulsifying properties of fractions of gluten peptides obtained by limited enzymatic hydrolysis and ultrafiltration. J. Cereal Sci. 2002, 35, 327–335. [Google Scholar] [CrossRef]
- Moro, A.; Báez, G.D.; Ballerini, G.A.; Busti, P.A.; Delorenzi, N.J. Emulsifying and foaming properties of β-lactoglobulin modified by heat treatment. Food Res. Int. 2013, 51, 1–7. [Google Scholar] [CrossRef]
- Bechaux, J.; Gatellier, P.; Le Page, J.F.; Drillet, Y.; Sante-Lhoutellier, V. A comprehensive review of bioactive peptides obtained from animal byproducts and their applications. Food Funct. 2019, 10, 6244–6266. [Google Scholar] [CrossRef] [PubMed]
- Drozłowska, E.; Weronis, M.; Bartkowiak, A. The influence of thermal hydrolysis process on emulsifying properties of potato protein isolate. J. Food Sci. Technol. 2020, 57, 1131–1137. [Google Scholar] [CrossRef] [PubMed]
- Benítez, R.; Ibarz, A.; Pagan, J. Hidrolizados de proteína: Procesos y aplicaciones. Acta Bioquim. Clin. Latinoam. 2008, 42, 227–236. [Google Scholar]
- Chew, L.Y.; Toh, G.T.; Ismail, A. Application of proteases for the production of bioactive peptides. In Enzymes in Food Biotechnology: Production, Applications, and Future Prospects; Kuddus, M., Ed.; Academic Press: London, UK, 2018; pp. 247–261. ISBN 9780128132807. [Google Scholar]
- Jeewanthi, R.K.C.; Lee, N.K.; Paik, H.D. Improved functional characteristics of whey protein hydrolysates in food industry. Korean J. Food Sci. Anim. Resour. 2015, 35, 350–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Padial-Domínguez, M.; Espejo-Carpio, F.J.; Pérez-Gálvez, R.; Guadix, A.; Guadix, E.M. Optimization of the emulsifying properties of food protein hydrolysates for the production of fish oil-in-water emulsions. Foods 2020, 9, 636. [Google Scholar] [CrossRef]
- Huang, X.L.; Catignani, G.L.; Swaisgood, H.E. Improved Emulsifying Properties of β-Barrel Domain Peptides Obtained by Membrane-Fractionation of a Limited Tryptic Hydrolysate of β-Lactoglobulin. J. Agric. Food Chem. 1996, 44, 3437–3443. [Google Scholar] [CrossRef]
- Turgeon, S.L.; Sanchez, C.; Gauthier, S.F.; Paquin, P. Stability and rheological properties of salad dressing containing peptidic fractions of whey proteins. Int. Dairy J. 1996, 6, 645–658. [Google Scholar] [CrossRef]
- Park, B.Y.; Yoon, K.Y. Functional properties of enzymatic hydrolysate and peptide fractions from perilla seed meal protein. Pol. J. Food Nutr. Sci. 2019, 69, 119–127. [Google Scholar] [CrossRef]
- Chabanon, G.; Chevalot, I.; Framboisier, X.; Chenu, S.; Marc, I. Hydrolysis of rapeseed protein isolates: Kinetics, characterization and functional properties of hydrolysates. Process Biochem. 2007, 42, 1419–1428. [Google Scholar] [CrossRef]
- Schmidt, M.M.; da Fontoura, A.M.; Vidal, A.R.; Dornelles, R.C.P.; Kubota, E.H.; de Mello, R.O.; Cansian, R.L.; Demiate, I.M.; de Oliveira, C.S. Characterization of hydrolysates of collagen from mechanically separated chicken meat residue. Food Sci. Technol. 2020, 40, 355–362. [Google Scholar] [CrossRef] [Green Version]
- Ling, Z.; Ai, M.; Zhou, Q.; Guo, S.; Zhou, L.; Fan, H.; Cao, Y.; Jiang, A. Fabrication egg white gel hydrolysates-stabilized oil-in-water emulsion and characterization of its stability and digestibility. Food Hydrocoll. 2020, 102, 105621. [Google Scholar] [CrossRef]
- Padial-Domínguez, M.; Espejo-Carpio, F.J.; García-Moreno, P.J.; Jacobsen, C.; Guadix, E.M. Protein derived emulsifiers with antioxidant activity for stabilization of omega-3 emulsions. Food Chem. 2020, 329, 127148. [Google Scholar] [CrossRef]
- Dai, L.; Hinrichs, J.; Weiss, J. Emulsifying properties of acid-hydrolyzed insoluble protein fraction from Chlorella protothecoides: Formation and storage stability of emulsions. Food Hydrocoll. 2020, 108, 105954. [Google Scholar] [CrossRef]
- Van der Ven, C.; Gruppen, H.; De Bont, D.B.A.; Voragen, A.G.J. Emulsion properties of casein and whey protein hydrolysates and the relation with other hydrolysate characteristics. J. Agric. Food Chem. 2001, 49, 5005–5012. [Google Scholar] [CrossRef]
- Caessens, P.W.J.R.; Visser, S.; Gruppen, H.; Voragen, A.G.J. β-Lactoglobulin hydrolysis. 1. Peptide composition and functional properties of hydrolysates obtained by the action of plasmin, trypsin, and Staphylococcus aureus V8 protease. J. Agric. Food Chem. 1999, 47, 2973–2979. [Google Scholar] [CrossRef]
- Schröder, A.; Berton-Carabin, C.; Venema, P.; Cornacchia, L. Interfacial properties of whey protein and whey protein hydrolysates and their influence on O/W emulsion stability. Food Hydrocoll. 2017, 73, 129–140. [Google Scholar] [CrossRef]
- Gao, Y.; Li, J.; Chang, C.; Wang, C.; Yang, Y.; Su, Y. Effect of enzymatic hydrolysis on heat stability and emulsifying properties of egg yolk. Food Hydrocoll. 2019, 97, 105224. [Google Scholar] [CrossRef]
- Bao, Z.J.; Zhao, Y.; Wang, X.Y.; Chi, Y.J. Effects of degree of hydrolysis (DH) on the functional properties of egg yolk hydrolysate with alcalase. J. Food Sci. Technol. 2017, 54, 669–678. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Lyu, F.; Zhou, X.; Wang, B.; Wang, X.; Ding, Y. Preparation of Skipjack Tuna (Katsuwonus pelamis) Protein Hydrolysate Using Combined Controlled Enzymatic Hydrolysis and Glycation for Improved Solubility and Emulsifying Properties. J. Food Nutr. Res. 2015, 3, 471–477. [Google Scholar] [CrossRef]
- Quan, T.H.; Benjakul, S. Production and characterisation of duck albumen hydrolysate using enzymatic process. Int. J. Food Sci. Technol. 2019, 54, 3015–3023. [Google Scholar] [CrossRef]
- Wu, W.U.; Hettiarachchy, N.S.; Qi, M. Hydrophobicity, solubility, and emulsifying properties of soy protein peptides prepared by papain modification and ultrafiltration. JAOCS J. Am. Oil Chem. Soc. 1998, 75, 845–850. [Google Scholar] [CrossRef]
- Kim, H.J.; Choi, S.J.; Shin, W.S.; Moon, T.W. Emulsifying properties of bovine serum albumin-galactomannan conjugates. J. Agric. Food Chem. 2003, 51, 1049–1056. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.; Yin, L.J.; Kobayashi, I.; Nakajima, M. Comparison of stability of bovine serum albumin-stabilized emulsions prepared by microchannel emulsification and homogenization. Food Hydrocoll. 2006, 20, 1020–1028. [Google Scholar] [CrossRef]
- Kiosseoglou, V.; Perdikis, A. Stability of bovine serum albumin-stabilized olive oil-in-water emulsions and the role of the oil minor surface-active lipids. Food Hydrocoll. 1994, 8, 27–32. [Google Scholar] [CrossRef]
- Saito, M.; Chikuni, K.; Monma, M.; Shimizu, M. Emulsifying and Oil-binding Properties of Bovine Serum Albumin and Its Enzymatic Hydrolyzate. Biosci. Biotechnol. Biochem. 1993, 57, 952–956. [Google Scholar] [CrossRef]
- Cheema, M.; Hristov, A.N.; Harte, F.M. The binding of orally dosed hydrophobic active pharmaceutical ingredients to casein micelles in milk. J. Dairy Sci. 2017, 100, 8670–8679. [Google Scholar] [CrossRef]
- Lorient, D.; Closs, B.; Courthaudon, J.L. Surface properties of the bovine casein components: Relationships between structure and foaming properties. J. Dairy Res. 1989, 56, 495–502. [Google Scholar] [CrossRef]
- Courthaudon, J.L.; Girardet, J.M.; Campagne, S.; Rouhier, L.M.; Campagna, S.; Linden, G.; Lorient, D. Surface active and emulsifying properties of casein micelles compared to those of sodium caseinate. Int. Dairy J. 1999, 9, 411–412. [Google Scholar] [CrossRef]
- Shimizu, M.; Lee, S.W.; Kaminogawa, S.; Yamauchi, K. Emulsifying Properties of an N-Terminal Peptide Obtained from the Peptic Hydrolyzate of αs1-Casein. J. Food Sci. 1984, 49, 1117–1120. [Google Scholar] [CrossRef]
- Zhai, J.; Miles, A.J.; Pattenden, L.K.; Lee, T.H.; Augustin, M.A.; Wallace, B.A.; Aguilar, M.I.; Wooster, T.J. Changes in β-lactoglobulin conformation at the oil/water interface of emulsions studied by synchrotron radiation circular dichroism spectroscopy. Biomacromolecules 2010, 11, 2136–2142. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.; Decker, E.A.; Xiao, H.; McClements, D.J. Comparison of biopolymer emulsifier performance in formation and stabilization of orange oil-in-water emulsions. JAOCS J. Am. Oil Chem. Soc. 2011, 88, 47–55. [Google Scholar] [CrossRef]
- Singh, H.; Sarkar, A. Behaviour of protein-stabilised emulsions under various physiological conditions. Adv. Colloid Interface Sci. 2011, 165, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Tu, M.; Cheng, S.; Lu, W.; Du, M. Advancement and prospects of bioinformatics analysis for studying bioactive peptides from food-derived protein: Sequence, structure, and functions. TrAC Trends Anal. Chem. 2018, 105, 7–17. [Google Scholar] [CrossRef]
- Rathinakumar, R.; Wimley, W.C. High-throughput discovery of broad-spectrum peptide antibiotics. FASEB J. 2010, 24, 3232–3238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuselier, T.; Wimley, W.C. Spontaneous Membrane Translocating Peptides: The Role of Leucine-Arginine Consensus Motifs. Biophys. J. 2017, 113, 835–846. [Google Scholar] [CrossRef]
- Selvaraj, C.; Sakkiah, S.; Tong, W.; Hong, H. Molecular dynamics simulations and applications in computational toxicology and nanotoxicology. Food Chem. Toxicol. 2018, 112, 495–506. [Google Scholar] [CrossRef]
- Iwaniak, A.; Darewicz, M.; Mogut, D.; Minkiewicz, P. Elucidation of the role of in silico methodologies in approaches to studying bioactive peptides derived from foods. J. Funct. Foods 2019, 61, 103486. [Google Scholar] [CrossRef]
- University of Warmia and Mazury in Olsztyn. BIOPEP-UWM. Available online: http://www.uwm.edu.pl/biochemia/index.php/en/biopep (accessed on 27 November 2020).
- Swiss Institute of Bioinformatics. PeptideCutter. Available online: https://web.expasy.org/peptide_cutter/ (accessed on 27 November 2020).
- Agyei, D.; Tsopmo, A.; Udenigwe, C.C. Bioinformatics and peptidomics approaches to the discovery and analysis of food-derived bioactive peptides. Anal. Bioanal. Chem. 2018, 410, 3463–3472. [Google Scholar] [CrossRef]
- Bioware. Available online: http://bioware.ucd.ie/~compass/biowareweb/ (accessed on 27 November 2020).
- Jahangiri, R.; Soltani, S.; Barzegar, A. A Review of QSAR Studies to Predict Activity of ACE Peptide Inhibitors. Pharm. Sci. 2014, 20, 122–129. [Google Scholar]
- Veerasamy, R.; Rajak, H.; Jain, A.; Sivadasan, S.; Varghese, C.P.; Agrawal, R.K. Validation of QSAR Models—Strategies and Importance. Int. J. Drug Des. Disocovery 2011, 2, 511–519. [Google Scholar]
- Ulmschneider, J.P.; Ulmschneider, M.B. Molecular Dynamics Simulations Are Redefining Our View of Peptides Interacting with Biological Membranes. Acc. Chem. Res. 2018, 51, 1106–1116. [Google Scholar] [CrossRef] [PubMed]
- Álvarez Vanegas, M.; Macías Lozano, A.; Núñez Vélez, V.; Garcés Ferreira, N.; Castro Barrera, H.; Álvarez Solano, O.; González Barrios, A.F. Molecular dynamics approach to investigate the coupling of the hydrophilic-lipophilic balance with the configuration distribution function in biosurfactant-based emulsions. J. Mol. Model. 2013, 19, 5539–5543. [Google Scholar] [CrossRef] [PubMed]
- She, A.Q.; Gang, H.Z.; Mu, B.Z. Temperature influence on the structure and interfacial properties of surfactin micelle: A molecular dynamics simulation study. J. Phys. Chem. B 2012, 116, 12735–12743. [Google Scholar] [CrossRef] [PubMed]
- Copps, J.; Murphy, R.F.; Lovas, S. Molecular dynamics simulations of peptides. Methods Mol. Biol. 2008, 494, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Huang, T.; Qiao, B.; Zhang, M.; Ma, H.; Zhang, L. Structures, dynamics, and water permeation free energy across bilayers of lipid a and its analog studied with molecular dynamics simulation. J. Phys. Chem. B 2014, 118, 13202–13209. [Google Scholar] [CrossRef]
- Chen, C.H.; Starr, C.G.; Troendle, E.; Wiedman, G.; Wimley, W.C.; Ulmschneider, J.P.; Ulmschneider, M.B. Simulation-Guided Rational de Novo Design of a Small Pore-Forming Antimicrobial Peptide. J. Am. Chem. Soc. 2019, 141, 4839–4848. [Google Scholar] [CrossRef]
- Farrotti, A.; Bocchinfuso, G.; Palleschi, A.; Rosato, N.; Salnikov, E.S.; Voievoda, N.; Bechinger, B.; Stella, L. Molecular dynamics methods to predict peptide locations in membranes: LAH4 as a stringent test case. Biochim. Biophys. Acta Biomembr. 2015, 1848, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Copps, J.; Murphy, R.F.; Lovas, S. Molecular Dynamics Simulations of Peptides. In Peptide-Based Drug Design. Methods in Molecular Biology; Otvos, L., Ed.; Humana Press: Totowa, NJ, USA, 2008; pp. 115–126. [Google Scholar]
- Rossi, G.; Fuchs, P.F.J.; Barnoud, J.; Monticelli, L. A coarse-grained MARTINI model of polyethylene glycol and of polyoxyethylene alkyl ether surfactants. J. Phys. Chem. B 2012, 116, 14353–14362. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, Y.; Tian, S.; Yan, H. Molecular Dynamics Simulation of Emulsification/Demulsification with a Gas Switchable Surfactant. J. Phys. Chem. C 2019, 123, 25246–25254. [Google Scholar] [CrossRef]
- Miyamoto, H.; Rein, D.M.; Ueda, K.; Yamane, C.; Cohen, Y. Molecular dynamics simulation of cellulose-coated oil-in-water emulsions. Cellulose 2017, 24, 2699–2711. [Google Scholar] [CrossRef]
- Aguilera-Segura, S.M.; Macias, A.; Carrero, D.; Vargas, W.; Vives-Florez, M.; Castro, H.; Alvarez, O.; Gonzalez, A. Escherichia coli´s OmpA as Biosurfactant for Cosmetic Industry: Stability Analysis and Experimental Validation Based on Molecular Simulations. In Advances in Computational Biology, Proceedings of the 2nd Colombian Congress on Computational Biology and Bioinformatics(CCBCOL); Castillo, F.L., Cristancho, M., Isaza, G., Pinzón, A., Rodríguez, C.J.M., Eds.; Springer International Publishing: Cham, Switzerland, 2014; pp. 265–271. [Google Scholar]
- Alting, A.C.; Pouvreau, L.; Giuseppin, M.L.F.; van Nieuwenhuijzen, N.H. Potato proteins. In Handbook of Food Proteins; Williams, P.A., Phillips, G.O., Eds.; Woodhead Publishing Limited: Cambridge, UK, 2011; pp. 316–334. [Google Scholar]
- Schmidt, J.M.; Damgaard, H.; Greve-Poulsen, M.; Larsen, L.B.; Hammershøj, M. Foam and emulsion properties of potato protein isolate and purified fractions. Food Hydrocoll. 2018, 74, 367–378. [Google Scholar] [CrossRef]
- Hanson, J.A.; Chang, C.B.; Graves, S.M.; Li, Z.; Mason, T.G.; Deming, T.J. Nanoscale double emulsions stabilized by single-component block copolypeptides. Nature 2008, 455, 85–88. [Google Scholar] [CrossRef]
- Wimley, W.C. Describing the Mechanism of Antimicrobial Peptide Action with the Interfacial Activity Model. ACS Chem. Biol. 2010, 5, 905–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castelletto, V.; Edwards-Gayle, C.J.C.; Hamley, I.W.; Barrett, G.; Seitsonen, J.; Ruokolainen, J. Peptide-Stabilized Emulsions and Gels from an Arginine-Rich Surfactant-like Peptide with Antimicrobial Activity. ACS Appl. Mater. Interfaces 2019, 11, 9893–9903. [Google Scholar] [CrossRef]
- Cantor, S.; Vargas, L.; Rojas, O.E.A.; Yarce, C.J.; Salamanca, C.H.; Oñate-Garzón, J. Evaluation of the antimicrobial activity of cationic peptides loaded in surface-modified nanoliposomes against foodborne bacteria. Int. J. Mol. Sci. 2019, 20, 680. [Google Scholar] [CrossRef] [Green Version]
- Fairman, R.; Chao, H.-G.; Mueller, L.; Lavoie, T.B.; Shen, L.; Novotny, J.; Matsueda, G.R. Characterization of a new four-chain coiled-coil: Influence of chain length on stability. Protein Sci. 1995, 4, 1457–1469. [Google Scholar] [CrossRef] [Green Version]
- Fukunishi, Y.; Tateishi, T.; Suzuki, M. Octane/water interfacial tension calculation by molecular dynamics simulation. J. Colloid Interface Sci. 1996, 180, 188–192. [Google Scholar] [CrossRef]
- Jones, D.B.; Middelberg, A.P.J. Mechanical properties of interfacially adsorbed peptide networks. Langmuir 2002, 18, 10357–10362. [Google Scholar] [CrossRef]
- Dexter, A.F.; Malcolm, A.S.; Middelberg, A.P.J. Reversible active switching of the mechanical properties of a peptide film at a fluid-fluid interface. Nat. Mater. 2006, 5, 502–506. [Google Scholar] [CrossRef] [PubMed]
- Malcolm, A.S.; Dexter, A.F.; Katakdhond, J.A.; Karakashev, S.I.; Nguyen, A.V.; Middelberg, A.P.J. Tuneable control of interfacial rheology and emulsion coalescence. ChemPhysChem 2009, 10, 778–781. [Google Scholar] [CrossRef] [PubMed]
- Dexter, A.F. Interfacial and emulsifying properties of designed β-strand peptides. Langmuir 2010, 26, 17997–18007. [Google Scholar] [CrossRef]
- Lab, H. Centre for Microbial Diseases and Immunity Research (CMDR). Available online: http://cmdr.ubc.ca/bobh/methods/methodsall.html (accessed on 15 April 2021).
- Yu, K.; Park, K.; Kim, Y.; Kang, S.W.; Shin, S.Y.; Hahm, K.S. Solution structure of a cathelicidin-derived antimicrobial peptide, CRAMP as determined by NMR spectroscopy. J. Pept. Res. 2002, 60, 1–9. [Google Scholar] [CrossRef]
- Wang, G.; Li, Y.; Li, X. Correlation of three-dimensional structures with the antibacterial activity of a group of peptides designed based on a nontoxic bacterial membrane anchor. J. Biol. Chem. 2005, 280, 5803–5811. [Google Scholar] [CrossRef] [Green Version]
- Mishra, B.; Wang, G. Ab Initio Design of Potent Anti-MRSA Peptides based on Database Filtering Technology. J. Chem. Soc. 2012, 134, 12426–12429. [Google Scholar] [CrossRef] [Green Version]
- Formulaction. Turbiscan Stability Index. Available online: https://www.formulaction.com/en/knowledge-center/turbiscan-stability-index (accessed on 8 February 2020).
- Menousek, J.; Mishra, B.; Hanke, M.L.; Heim, C.E.; Kielian, T.; Wang, G. Database screening and in vivo efficacy of antimicrobial peptides against methicillin-resistant Staphylococcus aureus USA300. Int. J. Antimicrob. Agents 2012, 39, 402–406. [Google Scholar] [CrossRef] [Green Version]
- Han, X.; Ning, W.; Ma, X.; Wang, X.; Zhou, K. Improving protein solubility and activity by introducing small peptide tags designed with machine learning models. Metab. Eng. Commun. 2020, 11, e00138. [Google Scholar] [CrossRef]
- Genchi, G.; Carocci, A.; Lauria, G.; Sinicropi, M.S.; Catalano, A. Nickel: Human health and environmental toxicology. Int. J. Environ. Res. Public Health 2020, 17, 679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czarnek, K.; Terpilowska, S.; Siwicki, A.K. Selected aspects of the action of cobalt ions in the human body. Cent. Eur. J. Immunol. 2015, 40, 236–242. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ricardo, F.; Pradilla, D.; Cruz, J.C.; Alvarez, O. Emerging Emulsifiers: Conceptual Basis for the Identification and Rational Design of Peptides with Surface Activity. Int. J. Mol. Sci. 2021, 22, 4615. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22094615
Ricardo F, Pradilla D, Cruz JC, Alvarez O. Emerging Emulsifiers: Conceptual Basis for the Identification and Rational Design of Peptides with Surface Activity. International Journal of Molecular Sciences. 2021; 22(9):4615. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22094615
Chicago/Turabian StyleRicardo, Fabian, Diego Pradilla, Juan C. Cruz, and Oscar Alvarez. 2021. "Emerging Emulsifiers: Conceptual Basis for the Identification and Rational Design of Peptides with Surface Activity" International Journal of Molecular Sciences 22, no. 9: 4615. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22094615