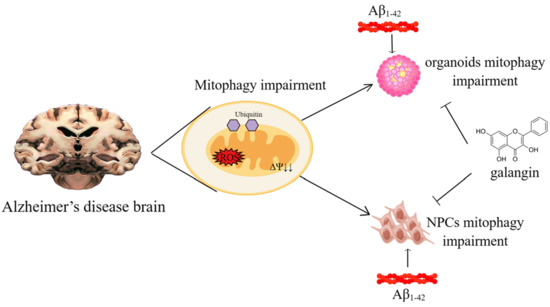
Galangin Rescues Alzheimer’s Amyloid-β Induced Mitophagy and Brain Organoid Growth Impairment
Abstract
:1. Introduction
2. Results
2.1. Aβ1-42 Suppressed the Growth of Brain Organoids and NPCs, as Well as Impaired Mitochondrial Function
2.2. Mitophagy Was Impaired by Aβ1-42 in Brain Organoid and NPCs
2.3. Galangin Rescued Aβ-Induced Mitophagy Impairment in Brain Organoids
2.4. Inhibition of Mitophagy Abolished the Effects of Galangin
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Compounds and Reagents
4.3. Aβ1-42 Oligomers Preparation
4.4. Human Brain Organoid Culture
4.5. EdU Detection
4.6. Cell Viability Assay
4.7. ROS Assay
4.8. Measurement of Mitochondrial Membrane Potential
4.9. Mitophagy Detection
4.10. Mitochondrial Content and Morphology Analysis
4.11. Western Blot
4.12. Immuostaining
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| NPCs | neural progenitor cells |
| MMP | mitochondrial membrane potential |
| ROS | reactive oxygen species |
| gal | galangin |
References
- Venkataraman, A.V.; Mansur, A.; Rizzo, G.; Bishop, C.; Lewis, Y.; Kocagoncu, E.; Lingford-Hughes, A.; Huiban, M.; Passchier, J.; Rowe, J.B.; et al. Widespread cell stress and mitochondrial dysfunction occur in patients with early Alzheimer’s disease. Sci. Transl. Med. 2022, 14, eabk1051. [Google Scholar] [CrossRef] [PubMed]
- Cen, X.; Zhang, M.; Zhou, M.; Ye, L.; Xia, H. Mitophagy regulates neurodegenerative diseases. Cells 2021, 10, 1876. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.T. Mechanisms of selective autophagy and mitophagy: Implications for neurodegenerative diseases. Neurobiol. Dis. 2019, 122, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Mary, A.; Eysert, F.; Checler, F.; Chami, M. Mitophagy in Alzheimer’s disease: Molecular defects and therapeutic approaches. Mol. Psychiatry 2022, 28, 202–216. [Google Scholar] [CrossRef] [PubMed]
- Fang, E.F.; Hou, Y.; Palikaras, K.; Adriaanse, B.A.; Kerr, J.S.; Yang, B.; Lautrup, S.; Hasan-Olive, M.M.; Caponio, D.; Dan, X.; et al. Mitophagy inhibits amyloid-beta and tau pathology and reverses cognitive deficits in models of Alzheimer’s disease. Nat. Neurosci. 2019, 22, 401–412. [Google Scholar] [CrossRef]
- Hu, Y.; Li, X.C.; Wang, Z.H.; Luo, Y.; Zhang, X.; Liu, X.P.; Feng, Q.; Wang, Q.; Yue, Z.; Chen, Z.; et al. Tau accumulation impairs mitophagy via increasing mitochondrial membrane potential and reducing mitochondrial parkin. Oncotarget 2016, 7, 17356–17368. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, T.; Ge, X.; Chen, J.; Zhao, Y.; Fu, J. Parkin overexpression attenuates abeta-induced mitochondrial dysfunction in HEK293 cells by restoring impaired mitophagy. Life Sci. 2020, 244, 117322. [Google Scholar] [CrossRef]
- Han, S.; Jeong, Y.Y.; Sheshadri, P.; Su, X.; Cai, Q. Mitophagy regulates integrity of mitochondria at synapses and is critical for synaptic maintenance. EMBO Rep. 2020, 21, e49801. [Google Scholar] [CrossRef]
- Xie, C.; Zhuang, X.X.; Niu, Z.; Ai, R.; Lautrup, S.; Zheng, S.; Jiang, Y.; Han, R.; Gupta, T.S.; Cao, S.; et al. Amelioration of Alzheimer’s disease pathology by mitophagy inducers identified via machine learning and a cross-species workflow. Nat. Biomed. Eng. 2022, 6, 76–93. [Google Scholar] [CrossRef]
- Cao, S.Q.; Aman, Y.; Fang, E.F.; Tencomnao, T.P. edulis extract protects against amyloid-beta toxicity in Alzheimer’s disease models through maintenance of mitochondrial homeostasis via the FOXO3/DAF-16 pathway. Mol. Neurobiol. 2022, 59, 5612–5629. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Knoblich, J.A. Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 2014, 9, 2329–2340. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Renner, M.; Martin, C.A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral organoids model human brain development and microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef]
- Clevers, H. Modeling development and disease with organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef]
- Kelava, I.; Lancaster, M.A. Stem cell models of human brain development. Cell Stem Cell 2016, 18, 736–748. [Google Scholar] [CrossRef]
- Pasca, A.M.; Sloan, S.A.; Clarke, L.E.; Tian, Y.; Makinson, C.D.; Huber, N.; Kim, C.H.; Park, J.Y.; O’Rourke, N.A.; Nguyen, K.D.; et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 2015, 12, 671–678. [Google Scholar] [CrossRef]
- Quadrato, G.; Nguyen, T.; Macosko, E.Z.; Sherwood, J.L.; Min Yang, S.; Berger, D.R.; Maria, N.; Scholvin, J.; Goldman, M.; Kinney, J.P.; et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature 2017, 545, 48–53. [Google Scholar] [CrossRef]
- Muguruma, K.; Nishiyama, A.; Kawakami, H.; Hashimoto, K.; Sasai, Y. Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep. 2015, 10, 537–550. [Google Scholar] [CrossRef]
- Yan, Y.; Song, L.; Bejoy, J.; Zhao, J.; Kanekiyo, T.; Bu, G.; Zhou, Y.; Li, Y. Modeling neurodegenerative microenvironment using cortical organoids derived from human stem cells. Tissue Eng. Part A 2018, 24, 1125–1137. [Google Scholar] [CrossRef]
- Bergmann, S.; Lawler, S.E.; Qu, Y.; Fadzen, C.M.; Wolfe, J.M.; Regan, M.S.; Pentelute, B.L.; Agar, N.Y.R.; Cho, C.F. Blood-brain-barrier organoids for investigating the permeability of CNS therapeutics. Nat. Protoc. 2018, 13, 2827–2843. [Google Scholar] [CrossRef]
- D’Avanzo, C.; Aronson, J.; Kim, Y.H.; Choi, S.H.; Tanzi, R.E.; Kim, D.Y. Alzheimer’s in 3D culture: Challenges and perspectives. BioEssays 2015, 37, 1139–1148. [Google Scholar] [CrossRef] [Green Version]
- Choi, S.H.; Kim, Y.H.; Quinti, L.; Tanzi, R.E.; Kim, D.Y. 3D culture models of Alzheimer’s disease: A road map to a “cure-in-a-dish”. Mol. Neurodegener. 2016, 11, 75. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, C.; Armijo, E.; Bravo-Alegria, J.; Becerra-Calixto, A.; Mays, C.E.; Soto, C. Modeling amyloid beta and tau pathology in human cerebral organoids. Mol. Psychiatry 2018, 23, 2363–2374. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Xu, R.; Wang, L.; Alam, M.M.; Ma, Z.; Zhu, S.; Martini, A.C.; Jadali, A.; Bernabucci, M.; Xie, P.; et al. Type-I-interferon signaling drives microglial dysfunction and senescence in human iPSC models of down syndrome and Alzheimer’s disease. Cell Stem Cell 2022, 29, 1135–1153. [Google Scholar] [CrossRef]
- Jan, A.; Hartley, D.M.; Lashuel, H.A. Preparation and characterization of toxic Aβ aggregates for structural and functional studies in Alzheimer’s disease research. Nat. Protoc. 2010, 5, 1186–1209. [Google Scholar] [CrossRef] [PubMed]
- Grimm, A.; Friedland, K.; Eckert, A. Mitochondrial dysfunction: The missing link between aging and sporadic Alzheimer’s disease. Biogerontology 2016, 17, 281–296. [Google Scholar] [CrossRef]
- Wang, X.; Su, B.; Perry, G.; Smith, M.A.; Zhu, X. Insights into amyloid-beta-induced mitochondrial dysfunction in Alzheimer disease. Free. Radic. Biol. Med. 2007, 43, 1569–1573. [Google Scholar] [CrossRef]
- Holland, T.M.; Agarwal, P.; Wang, Y.; Leurgans, S.E.; Bennett, D.A.; Booth, S.L.; Morris, M.C. Dietary flavonols and risk of Alzheimer dementia. Neurology 2020, 94, e1749–e1756. [Google Scholar] [CrossRef]
- Aloud, A.A.; Veeramani, C.; Govindasamy, C.; Alsaif, M.A.; El Newehy, A.S.; Al-Numair, K.S. Galangin, a dietary flavonoid, improves antioxidant status and reduces hyperglycemia-mediated oxidative stress in streptozotocin-induced diabetic rats. Redox Rep. 2017, 22, 290–300. [Google Scholar] [CrossRef]
- Das, S.; Majumder, T.; Sarkar, A.; Mukherjee, P.; Basu, S. Flavonoids as BACE1 inhibitors: QSAR modelling, screening and in vitro evaluation. Int. J. Biol. Macromol. 2020, 165, 1323–1330. [Google Scholar] [CrossRef]
- Huang, L.; Lin, M.; Zhong, X.; Yang, H.; Deng, M. Galangin decreases p-tau, Abeta42 and beta-secretase levels, and suppresses autophagy in okadaic acid-induced PC12 cells via an Akt/GSK3beta/mTOR signaling-dependent mechanism. Mol. Med. Rep. 2019, 19, 1767–1774. [Google Scholar]
- Zeng, H.; Huang, P.; Wang, X.; Wu, J.; Wu, M.; Huang, J. Galangin-induced down-regulation of BACE1 by epigenetic mechanisms in SH-SY5Y cells. Neuroscience 2015, 294, 172–181. [Google Scholar] [CrossRef]
- Choi, M.J.; Lee, E.J.; Park, J.S.; Kim, S.N.; Park, E.M.; Kim, H.S. Anti-inflammatory mechanism of galangin in lipopolysaccharide-stimulated microglia: Critical role of PPAR-gamma signaling pathway. Biochem. Pharmacol. 2017, 144, 120–131. [Google Scholar] [CrossRef]
- Dagda, R.K.; Cherra, S.J., 3rd; Kulich, S.M.; Tandon, A.; Park, D.; Chu, C.T. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J. Biol. Chem. 2009, 284, 13843–13855. [Google Scholar] [CrossRef] [Green Version]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, R.; Lu, J.; Pei, G.; Huang, S. Galangin Rescues Alzheimer’s Amyloid-β Induced Mitophagy and Brain Organoid Growth Impairment. Int. J. Mol. Sci. 2023, 24, 3398. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms24043398
Zhang R, Lu J, Pei G, Huang S. Galangin Rescues Alzheimer’s Amyloid-β Induced Mitophagy and Brain Organoid Growth Impairment. International Journal of Molecular Sciences. 2023; 24(4):3398. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms24043398
Chicago/Turabian StyleZhang, Ru, Juan Lu, Gang Pei, and Shichao Huang. 2023. "Galangin Rescues Alzheimer’s Amyloid-β Induced Mitophagy and Brain Organoid Growth Impairment" International Journal of Molecular Sciences 24, no. 4: 3398. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms24043398