Cryptosporidium Oocyst Contamination in Drinking Water: A Case Study in Italy
Abstract
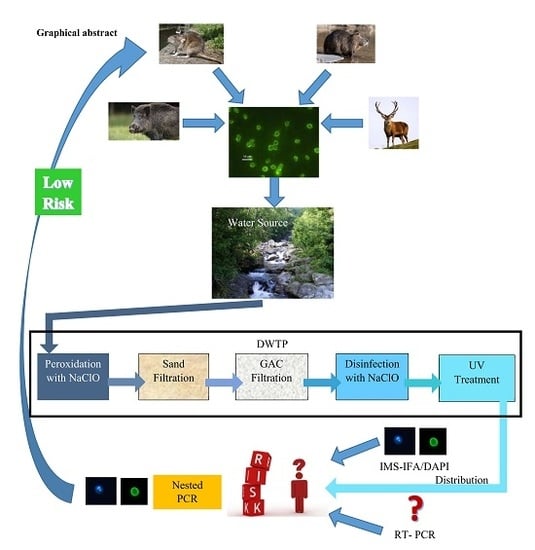
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Drinking Water Treatment Plant
2.3. Cryptosporidium Detection by IFA/DAPI
- Recovery% = 100 × [(Nsp − Nusp)/T]
- Nsp = number of oocysts counted in the spiked sample
- Nusp = number of oocysts counted in the unspiked sample
- T = true value of the oocysts or cysts spiked
- The oocyst concentration was calculated with the formula reported below:
- N oocysts/100 L = [(Nm × 2)/Ls] × 100
- Nm = number of oocysts determined by microscopy
- Ls = number of liters sampled
- Viability % = (Nn × 100)/No
- Nn = number of oocysts with DAPI-positive nuclei
- No = number of IFA-positive oocysts
2.4. Cryptosporidium Detection by Reverse Transcriptase-PCR (RT-PCR)
2.5. Cryptosporidium spp. PCR
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Benedict, K.M.; Reses, H.; Vigar, M.; Roth, D.M.; Roberts, V.A.; Mattioli, M.; Cooley, L.A.; Hilborn, E.D.; Wade, T.J.; Fullerton, K.E.; et al. Surveillance for Waterborne Disease Outbreaks Associated with Drinking Water—United States, 2013–2014. MMWR 2017, 10, 1216–1221. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority and European Centre for Disease Prevention and Control (EFSA and ECDC). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 2018, 16, e05500. [Google Scholar]
- Efstratiou, A.; Ongerth, J.E.; Karanis, P. Waterborne transmission of protozoan parasites: Review of worldwide outbreaks—An update 2011–2016. Water Res. 2017, 114, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Zahedi, A.; Monis, P.; Gofton, A.W.; Oskam, C.L.; Ball, A.; Bath, A.; Bartkow, M.; Robertson, I.; Ryan, U. Cryptosporidium species and subtypes in animals inhabiting drinking water catchments in three states across Australia. Water Res. 2018, 134, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Cacciò, S.M.; Putignani, L. Epidemiology of human cryptosporidiosis. In Cryptosporidium: Parasite and Disease; Cacciò, S.M., Widmer, G., Eds.; Springer: Vienna, Austria, 2014; pp. 43–79. [Google Scholar]
- Bonesa, A.J.; Josséa, L.; Morea, C.; Millera, C.N.; Michaelis, M.; Tsaousisa, A.D. Past and future trends of Cryptosporidium in vitro research. Exp. Parasitol. 2019, 196, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Corso, P.S.; Kramer, M.H.; Blair, K.A.; Addiss, D.G.; Davis, J.P.; Haddix, A.C. Cost of Illness in the 1993 Waterborne Cryptosporidium Outbreak, Milwaukee, Wisconsin. Emerg. Infect. Dis. 2003, 9, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, R.M. Waterborne outbreaks of cryptosporidiosis. Annali dell’Istituto superiore di sanita 2012, 48, 429–446. [Google Scholar] [CrossRef] [PubMed]
- Rochelle, P.A.; Di Giovanni, G.D. Cryptosporidium oocysts in drinking water and recreational water. In Cryptosporidium: Parasite and Disease; Cacciò, S.M., Widmer, G., Eds.; Springer: Vienna, Austria, 2014; pp. 489–513. [Google Scholar]
- Robertson, L.J.; Chalmers, R.M. Foodborne cryptosporidiosis: Is there really more in Nordic countries? Trends Parasitol. 2013, 29, 3–9. [Google Scholar] [CrossRef] [PubMed]
- McLauchlin, J.; Amar, C.; Pedraza-Díaz, S.; Nichols, G.L. Molecular epidemiological analysis of Cryptosporidium spp. in the United Kingdom: Results of genotyping Cryptosporidium spp. in 1705 fecal samples from humans and 105 fecal samples from livestock animals. J. Clin. Microbiol. 2000, 38, 3984–3990. [Google Scholar] [PubMed]
- Lake, I.R.; Nichols, G.; Bentham, G.; Harrison, F.C.; Hunter, P.R.; Kovats, S.R. Cryptosporidiosis decline after regulation, England and Wales, 1989–2005. Emerg. Infect. Dis. 2007, 13, 623–625. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, R.M.; Elwin, K.; Thomas, A.L.; Guy, E.C.; Mason, B. Long-term Cryptosporidium typing reveals the aetiology and species-specific epidemiology of human cryptosporidiosis in England and Wales, 2000 to 2003. Euro Surveill. 2009, 14, 19086. [Google Scholar] [CrossRef] [PubMed]
- Rehn, M.; Wallensten, A.; Widerström, M.; Lilja, M.; Grunewald, M.; Stenmark, S.; Kark, M.; Lindh, J. Post-infection symptoms following two large waterborne outbreaks of Cryptosporidium hominis in Northern Sweden, 2010–2011. BMC Public Health 2015, 15, 529. [Google Scholar] [CrossRef] [PubMed]
- Widerström, M.; Schönning, C.; Lilja, M.; Lebbad, M.; Ljung, T.; Allestam, G.; Ferm, M.; Björkholm, B.; Hansen, A.; Hiltula, J.; et al. Large outbreak of Cryptosporidium hominis infection transmitted through the public water supply, Sweden. Emerg. Infect. Dis. 2014, 20, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Pozio, E.; Rezza, G.; Boschini, A.; Pezzotti, P.; Tamburrini, A.; Rossi, P.; Di Fine, M.; Smacchia, C.; Schiesari, A.; Gattei, E.; et al. Clinical cryptosporidiosis and human immunodeficiency virus (HIV)-induced immunosuppression: Findings from a longitudinal study of HIV-positive and HIV-negative former injection drug users. J. Infect. Dis. 1997, 176, 969–975. [Google Scholar] [CrossRef] [PubMed]
- Cacciò, S.M.; Chalmers, R.M. Human cryptosporidiosis in Europe. Clin. Microbiol. Infect. 2016, 22, 471–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betancourt, W.Q.; Rose, J.B. Drinking water treatment processes for removal of Cryptosporidium and Giardia. Vet. Parasitol. 2004, 126, 219–234. [Google Scholar] [CrossRef] [PubMed]
- Rousseau, A.; La Carbona, S.; Dumetre, A.; Robertson, L.J.; Gargala, G.; Escotte-Binet, S.; Favennec, L.; Villena, I.; Gerard, C.; Aubert, D. Assessing viability and infectivity of foodborne and waterborne stages (cysts/oocysts) of Giardia duodenalis, Cryptosporidium spp., and Toxoplasma gondii: A review of methods. Parasite 2018, 25, 14. [Google Scholar] [CrossRef] [PubMed]
- ISS. Metodi analitici per le acque destinate al consumo umano ai sensi del D. lgs. 31/2001. Metodi microbiologici. A cura di Lucia Bonadonna e Massimo Ottaviani; Rapporti ISTISAN: Roma, Italy, 2007; ISSN 1123-3117. [Google Scholar]
- Kaucner, C.; Stinear, T. Sensitive and rapid detection of viable Giardia cysts and Cryptosporidium parvum oocysts in large-volume water samples with wound fiberglass cartridge filters and reverse transcription-PCR. Appl. Environ. Microbiol. 1998, 64, 1743–1749. [Google Scholar]
- Ryan, U.; Xiao, L.; Read, C.; Zhou, L.; Lal, A.A.; Pavlasek, I. Identification of novel Cryptosporidium genotypes from the Czech Republic. Appl. Environ. Microb. 2003, 69, 4302–4307. [Google Scholar] [CrossRef]
- Amoah, K.; Craik, S.; Smith, D.W.; Belosevic, M. Inactivation of Cryptosporidium oocysts and Giardia cysts by ultraviolet light in the presence of natural particulate matter. J. Water Supply Res. Technol. 2005, 54, 165–178. [Google Scholar] [CrossRef]
- Kar, S.; Guven, E.; Karaer, Z. Effects of Laminar Flow Ultraviolet Sterilization Protocol on Cryptosporidium parvum oocysts and determination of these effects with vital dyes. Kafkas Univ. Vet. Fak. Derg. 2011, 17, 21–24. [Google Scholar]
- Vande Burgt, N.H.; Auer, A.; Zintl, A. Comparison of in vitro viability methods for Cryptosporidium oocysts. Exp. Parasitol. 2018, 187, 30–36. [Google Scholar] [CrossRef] [PubMed]
- EPA. Method 1623: Cryptosporidium and Giardia in Water by Filtration/IMS/FA. 2005. Available online: https://www.epa.gov/sites/production/files/2015-07/documents/epa-1623.pdf (accessed on 30 April 2019).
- Stokdyk, J.P.; Spencer, S.K.; Walsh, J.F.; de Lamber, J.R.; Firnstahl, A.D.; Anderson, A.C.; Rezania, L.I.W.; Borchardt, M.A. Cryptosporidium incidence and surface water influence of groundwater supplying public water systems in Minnesota, USA. Environ. Sci. Technol. 2019, 53, 3391–3398. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, R.M.; Robinson, G.; Elwin, K.; Elson, R. Analysis of the Cryptosporidium spp. and gp60 subtypes linked to human outbreaks of cryptosporidiosis in England and Wales, 2009 to 2017. Parasites Vectors 2019, 12, 95. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, C.; López, M.C.; Galeano, L.A.; Qvarnstrom, Y.; Houghton, K.; Ramírez, J.D. Molecular detection and genotyping of pathogenic protozoan parasites in raw and treated water samples from southwest Colombia. Parasites Vectors 2018, 11, 563. [Google Scholar] [CrossRef] [PubMed]
- Puleston, R.L.; Mallaghan, C.M.; Modha, D.E.; Hunter, P.R.; Nguyen-Van-Tam, J.S.; Regan, C.M.; Nichols, G.L.; Chalmers, R.M. The first recorded outbreak of cryptosporidiosis due to Cryptosporidium cuniculus (formerly rabbit genotype), following a water quality incident. J. Water Health 2014, 12, 41–50. [Google Scholar] [CrossRef] [PubMed]

| Date | Sampling Point | Liters Filtered | FITC Oocysts/100 L a | DAPI Oocysts/100 L b | Viability % c |
|---|---|---|---|---|---|
| May, 2013 | OUT DWTP | 352 | ND | ND | NC |
| Tap Water | 568 | ND | ND | NC | |
| June, 2013 | OUT DWTP | 480 | ND | ND | NC |
| Tap Water | 742 | ND | ND | NC | |
| August, 2013 | IN DWTP | 303 | 1.98 | ND | NC |
| OUT DWTP | 294 | 0.68 | ND | NC | |
| Tap Water | 372 | 20.43 | 19.89 | 97.36 | |
| October, 2013 | IN DWTP | 252 | 12.7 | 3.17 | 24.96 |
| OUT DWTP | 375 | 13.87 | 11.74 | 84.64 | |
| Tap Water | 402 | 1.49 | 0.50 | 33.39 | |
| December, 2013 | OUT DWTP | 300 | 2.00 | 0.67 | 33.50 |
| Tap Water | 375 | 1.07 | ND | NC | |
| March, 2014 | IN DWTP | 268 | 4.48 | 2.24 | 50.00 |
| OUT DWTP | 391 | 1.53 | 0.51 | 33.33 | |
| Tap Water | 455 | 1.76 | 0.88 | 50.00 | |
| April, 2014 | IN DWTP | 296 | 2.03 | 0.68 | 33.50 |
| OUT DWTP | 324 | 2.47 | 1.23 | 49.80 | |
| Tap Water | 584 | 1.37 | 1.37 | 100.00 | |
| July, 2015 | IN DWTP | 413 | 1.45 | 1.45 | 100.00 |
| OUT DWTP | 382 | 0.26 | 0.26 | 100.00 | |
| Tap Water | 1,500 | 0.33 | 0.33 | 100.00 | |
| May, 2016 | IN DWTP | 155 | 11.29 | 11.29 | 100.00 |
| OUT DWTP | 610 | 7.21 | 6.23 | 86.41 | |
| Tap Water | 810 | 15.06 | 12.10 | 80.34 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pignata, C.; Bonetta, S.; Bonetta, S.; Cacciò, S.M.; Sannella, A.R.; Gilli, G.; Carraro, E. Cryptosporidium Oocyst Contamination in Drinking Water: A Case Study in Italy. Int. J. Environ. Res. Public Health 2019, 16, 2055. https://0-doi-org.brum.beds.ac.uk/10.3390/ijerph16112055
Pignata C, Bonetta S, Bonetta S, Cacciò SM, Sannella AR, Gilli G, Carraro E. Cryptosporidium Oocyst Contamination in Drinking Water: A Case Study in Italy. International Journal of Environmental Research and Public Health. 2019; 16(11):2055. https://0-doi-org.brum.beds.ac.uk/10.3390/ijerph16112055
Chicago/Turabian StylePignata, Cristina, Silvia Bonetta, Sara Bonetta, Simone M. Cacciò, Anna R. Sannella, Giorgio Gilli, and Elisabetta Carraro. 2019. "Cryptosporidium Oocyst Contamination in Drinking Water: A Case Study in Italy" International Journal of Environmental Research and Public Health 16, no. 11: 2055. https://0-doi-org.brum.beds.ac.uk/10.3390/ijerph16112055