Hyperspectral Dimensionality Reduction Based on Multiscale Superpixelwise Kernel Principal Component Analysis
Abstract
:1. Introduction
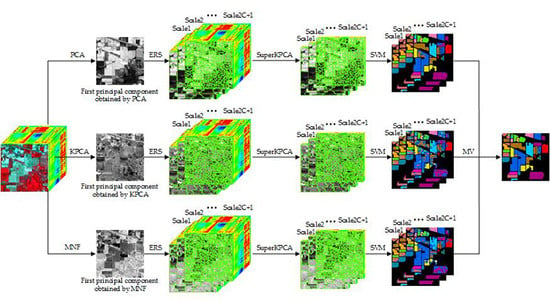
2. Proposed Methods
2.1. Multiscale Superpixel Segmentation
2.2. Kernel Principal Component Analysis
2.3. Classification and Fusion
| Algorithm 1: Proposed MSuperKPCA for an HSI |
| 1. INPUT: (1) data: a hyperspectral image and its training sample set and testing sample set (2) parameters: the reduced dimensions , the number of fundamental superpixels , the segmentation scale |
| 2. OUTPUT: a 2-D classification map |
| 3. Begin |
| 4. calculate the number of superpixels at each scale: |
| 5. convert all values of to decimals: |
| 6. use PCA to get the first PC of and convert it to unit format |
| 7. for each number of superpixels in do |
| 8. (1) use ERS to segment into corresponding numbers of superpixels |
| 9. (2) perform KPCA on the hyperspectral data in each superpixel and take the first PCs |
| 10. (3) combine the dimensionality reduction results in each superpixel and get |
| 11. (4) use SVM to classify the dimensionality reduction result |
| 12. end for |
| 13. for each pixel in the hyperspectral image do |
| 14. take the category with the most occurrences among candidate categories as the final classification result |
| 15. end for |
| 14. End |
3. Experiments and Results
3.1. Datasets Description and Experimental Setting
3.2. Parameters Settings
3.2.1. Analysis of the Influence of the Number of Superpixels
3.2.2. Analysis of the Impact of the Segmentation Scale
3.3. Fusion of Multiscale Classification Results Based on Different Fundamental Images
4. Discussion
4.1. Comparison with the State-of-the-art Approaches
4.2. Running Times
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Li, W.; Feng, F.; Li, H.; Du, Q. Discriminant analysis-based dimension reduction for hyperspectral image classification: A survey of the most recent advances and an experimental comparison of different techniques. IEEE Geosci. Remote Sens. Mag. 2018, 6, 15–34. [Google Scholar] [CrossRef]
- Hughes, G. On the mean accuracy of statistical pattern recognizers. IEEE Trans. Inf. Theory 1968, 14, 55–63. [Google Scholar] [CrossRef]
- Pearson, K. LIII. On lines and planes of closest fit to systems of points in space. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1901, 2, 559–572. [Google Scholar] [CrossRef]
- Bachmann, C.M.; Ainsworth, T.L.; Fusina, R.A. Exploiting manifold geometry in hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 2005, 43, 441–454. [Google Scholar] [CrossRef]
- Keshava, N.; Mustard, J.F. Spectral unmixing. IEEE Signal Process. Mag. 2002, 19, 44–57. [Google Scholar] [CrossRef]
- Scholz, M.; Kaplan, F.; Guy, C.L.; Kopka, J.; Selbig, J. Non-linear PCA: A missing data approach. Bioinformatics 2005, 21, 3887–3895. [Google Scholar] [CrossRef]
- Zhang, H.; Pedrycz, W.; Miao, D.; Wei, Z. From principal curves to granular principal curves. IEEE Trans. Cybern. 2014, 44, 748–760. [Google Scholar] [CrossRef]
- Lu, H.; Plataniotis, K.N.; Venetsanopoulos, A.N. A survey of multilinear subspace learning for tensor data. Pattern Recognit. 2011, 44, 1540–1551. [Google Scholar] [CrossRef]
- Schölkopf, B.; Smola, A.; Müller, K.-R. Nonlinear component analysis as a kernel eigenvalue problem. Neural Comput. 1998, 10, 1299–1319. [Google Scholar] [CrossRef]
- Filho, J.B.O.S.; Diniz, P.S.R. A fixed-point online kernel principal component extraction algorithm. IEEE Trans. Signal Process. 2017, 65, 6244–6259. [Google Scholar] [CrossRef]
- Filho, J.B.O.S.; Diniz, P.S.R. Improving KPCA online extraction by orthonormalization in the feature space. IEEE Trans. Neural Netw. Learn. Syst. 2018, 29, 1382–1387. [Google Scholar] [CrossRef] [PubMed]
- Washizawa, Y. Adaptive subset kernel principal component analysis for time-varying patterns. IEEE Trans. Neural Netw. Learn. Syst. 2012, 23, 1961–1973. [Google Scholar] [CrossRef] [PubMed]
- Gan, L.; Xia, J.; Du, P.; Chanussot, J. Multiple feature kernel sparse representation classifier for hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 2018, 56, 5343–5356. [Google Scholar] [CrossRef]
- Honeine, P. Online kernel principal component analysis: A reduced-order model. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 1814–1826. [Google Scholar] [CrossRef] [PubMed]
- Papaioannou, A.; Zafeiriou, S. Principal component analysis with complex kernel: The widely linear Model. IEEE Trans. Neural Netw. Learn. Syst. 2014, 25, 1719–1726. [Google Scholar] [CrossRef]
- Chakour, C.; Benyounes, A.; Boudiaf, M. Diagnosis of uncertain nonlinear systems using interval kernel principal components analysis: Application to a weather station. ISA Trans. 2018, 83, 126–141. [Google Scholar] [CrossRef]
- Jaffel, I.; Taouali, O.; Harkat, M.F.; Messaoud, H. Moving window KPCA with reduced complexity for nonlinear dynamic process monitoring. I|Sa Trans. 2016, 64, 184–192. [Google Scholar] [CrossRef]
- Jia, S.; Wu, K.; Zhu, J.; Jia, X. Spectral–spatial gabor surface feature fusion Approach for hyperspectral imagery classification. IEEE Trans. Geosci. Remote Sens. 2019, 57, 1142–1154. [Google Scholar] [CrossRef]
- Guo, Y.; Jiao, L.; Wang, S.; Wang, S.; Liu, F.; Hua, W. Fuzzy superpixels for polarimetric SAR images classification. IEEE Trans. Fuzzy Syst. 2018, 26, 2846–2860. [Google Scholar] [CrossRef]
- Berveglieri, A.; Imai, N.N.; Tommaselli, A.M.G.; Casagrande, B.; Honkavaara, E. Successional stages and their evolution in tropical forests using multi-temporal photogrammetric surface models and superpixels. ISPRS J. Photogramm. Remote Sens. 2018, 146, 548–558. [Google Scholar] [CrossRef]
- Huang, X.; Chen, H.; Gong, J. Angular difference feature extraction for urban scene classification using ZY-3 multi-angle high-resolution satellite imagery. ISPRS J. Photogramm. Remote Sens. 2018, 135, 127–141. [Google Scholar] [CrossRef]
- Jia, S.; Deng, B.; Zhu, J.; Jia, X.; Li, Q. Local binary pattern-based hyperspectral image classification with superpixel guidance. IEEE Trans. Geosci. Remote Sens. 2018, 56, 749–759. [Google Scholar] [CrossRef]
- Belgiu, M.; Csillik, O. Sentinel-2 cropland mapping using pixel-based and object-based time-weighted dynamic time warping analysis. Remote Sens. Environ. 2018, 204, 509–523. [Google Scholar] [CrossRef]
- Zhang, C.; Sargent, I.; Pan, X.; Li, H.; Gardiner, A.; Hare, J.; Atkinson, P.M. Joint deep learning for land cover and land use classification. Remote Sens. Environ. 2019, 221, 173–187. [Google Scholar] [CrossRef]
- Zhang, C.; Sargent, I.; Pan, X.; Li, H.; Gardiner, A.; Hare, J.; Atkinson, P.M. An object-based convolutional neural network (OCNN) for urban land use classification. Remote Sens. Environ. 2018, 216, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Massey, R.; Sankey, T.T.; Yadav, K.; Congalton, R.G.; Tilton, J.C. Integrating cloud-based workflows in continental-scale cropland extent classification. Remote Sens. Environ. 2018, 219, 162–179. [Google Scholar] [CrossRef]
- Fauvel, M.; Tarabalka, Y.; Benediktsson, J.A.; Chanussot, J.; Tilton, J.C. Advances in spectral-spatial classification of hyperspectral images. Proc. IEEE 2013, 101, 652–675. [Google Scholar] [CrossRef]
- Lv, X.; Ming, D.; Lu, T.; Zhou, K.; Wang, M.; Bao, H. A new method for region-based majority voting CNNs for very high resolution image classification. Remote Sens. 2018, 10, 1946. [Google Scholar] [CrossRef]
- Feng, J.; Wang, L.; Yu, H.; Jiao, L.; Zhang, X. Divide-and-conquer dual-architecture convolutional neural network for classification of hyperspectral images. Remote Sens. 2019, 11, 484. [Google Scholar] [CrossRef]
- Zhang, S.; Li, S.; Fu, W.; Fang, L. Multiscale superpixel-based sparse representation for hyperspectral Image classification. Remote Sens. 2017, 9, 139. [Google Scholar] [CrossRef]
- Cui, B.; Xie, X.; Ma, X.; Ren, G.; Ma, Y. Superpixel-based extended random walker for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2018, 56, 3233–3243. [Google Scholar] [CrossRef]
- Jin, X.; Gu, Y. Superpixel-based intrinsic image decomposition of hyperspectral images. IEEE Trans. Geosci. Remote Sens. 2017, 55, 4285–4295. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, J.; Chen, C.; Wang, Z.; Cai, Z.; Wang, L. Super PCA: A superpixelwise PCA approach for unsupervised feature extraction of hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 2018, 56, 4581–4593. [Google Scholar] [CrossRef]
- Mountrakis, G.; Im, J.; Ogole, C. Support vector machines in remote sensing: A review. ISPRS J. Photogramm. Remote Sens. 2011, 66, 247–259. [Google Scholar] [CrossRef]
- Mohammadimanesh, F.; Salehi, B.; Mahdianpari, M.; Motagh, M.; Brisco, B. An efficient feature optimization for wetland mapping by synergistic use of SAR intensity, interferometry, and polarimetry data. Int. J. Appl. Earth Obs. Geoinf. 2018, 73, 450–462. [Google Scholar] [CrossRef]
- Green, A.A.; Berman, M.; Switzer, P.; Craig, M.D. A transformation for ordering multispectral data in terms of image quality with implications for noise removal. IEEE Trans. Geosci. Remote Sens. 1988, 26, 65–74. [Google Scholar] [CrossRef] [Green Version]
- Achanta, R.; Shaji, A.; Smith, K.; Lucchi, A.; Fua, P.; Süsstrunk, S. SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 2274–2282. [Google Scholar] [CrossRef] [PubMed]
- Vincent, L.; Soille, P. Watersheds in digital spaces: An efficient algorithm based on immersion simulations. IEEE Trans. Pattern Anal. Mach. Intell. 1991, 13, 583–598. [Google Scholar] [CrossRef]
- Comaniciu, D.; Meer, P. Mean shift: A robust approach toward feature space analysis. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 603–619. [Google Scholar] [CrossRef]
- Liu, M.; Tuzel, O.; Ramalingam, S.; Chellappa, R. Entropy rate superpixel segmentation. In Proceedings of the CVPR 2011, Colorado Springs, CO, USA, 20–25 June 2011; pp. 2097–2104. [Google Scholar]
- Dong, X.; Shen, J.; Shao, L.; Gool, L.V. Sub-markov random walk for image segmentation. IEEE Trans. Image Process. 2016, 25, 516–527. [Google Scholar] [CrossRef]
- Felzenszwalb, P.F.; Huttenlocher, D.P. Efficient graph-based image segmentation. Int. J. Comput. Vis. 2004, 59, 167–181. [Google Scholar] [CrossRef]
- Jianbo, S.; Malik, J. Normalized cuts and image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2000, 22, 888–905. [Google Scholar] [CrossRef]
- Liang, X.; Xu, C.; Shen, X.; Yang, J.; Tang, J.; Lin, L.; Yan, S. Human parsing with contextualized convolutional neural network. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 115–127. [Google Scholar] [CrossRef]
- He, Z.; Liu, L.; Zhou, S.; Shen, Y. Learning group-based sparse and low-rank representation for hyperspectral image classification. Pattern Recognit. 2016, 60, 1041–1056. [Google Scholar] [CrossRef]
- Cao, L.; Luo, F.; Chen, L.; Sheng, Y.; Wang, H.; Wang, C.; Ji, R. Weakly supervised vehicle detection in satellite images via multi-instance discriminative learning. Pattern Recognit. 2017, 64, 417–424. [Google Scholar] [CrossRef]
- Widjaja, D.; Varon, C.; Dorado, A.; Suykens, J.A.K.; Huffel, S.V. Application of kernel principal component analysis for single-lead-ECG-derived respiration. IEEE Trans. Biomed. Eng. 2012, 59, 1169–1176. [Google Scholar] [CrossRef]
- Xie, L.; Li, Z.; Zeng, J.; Kruger, U. Block adaptive kernel principal component analysis for nonlinear process monitoring. AIChE J. 2016, 62, 4334–4345. [Google Scholar] [CrossRef]
- Chang, C.-C.; Lin, C.-J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 1–27. [Google Scholar] [CrossRef]














| Indian Pines | Pavia University | Salinas | ||||
|---|---|---|---|---|---|---|
| Class | Categories | Samples | Categories | Samples | Categories | Samples |
| 1 | alfalfa | 46 | asphalt | 6631 | weeds_1 | 2009 |
| 2 | corn-no till | 1428 | meadows | 18,649 | weeds_2 | 3726 |
| 3 | corn-min | 830 | gravel | 2099 | fallow | 1976 |
| 4 | corn | 237 | trees | 3064 | fallow plow | 1394 |
| 5 | grass or pasture | 483 | metal sheets | 1345 | fallow smooth | 2678 |
| 6 | grass or trees | 730 | bare boil | 5029 | stubble | 3959 |
| 7 | grass or pasture-mowed | 28 | bitumen | 1330 | celery | 3579 |
| 8 | hay-windrowed | 478 | bricks | 3682 | grapes | 11,271 |
| 9 | oats | 20 | shadows | 947 | soil | 6203 |
| 10 | soybean-no till | 972 | corn | 3278 | ||
| 11 | soybean-min | 2455 | lettuce 4 wk | 1068 | ||
| 12 | soybean-clean | 593 | lettuce 5 wk | 1927 | ||
| 13 | wheat | 205 | lettuce 6 wk | 916 | ||
| 14 | woods | 1265 | lettuce 7 wk | 1070 | ||
| 15 | buildings-grass-tree-drives | 386 | vineyard untrained | 7268 | ||
| 16 | stone-steel towers | 93 | vineyard trellis | 1807 | ||
| total | 10,249 | total | 42,776 | total | 54,129 | |
| Indian Pines | Pavia University | Salinas | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | PCA | KPCA | MNF | 3M 1 | PCA | KPCA | MNF | 3M | PCA | KPCA | MNF | 3M |
| 1 | 97.56 | 97.56 | 97.56 | 97.56 | 75.42 | 76.37 | 84.38 | 83.76 | 100.00 | 100.00 | 100.00 | 100.00 |
| 2 | 77.30 | 68.59 | 84.68 | 72.24 | 94.07 | 89.83 | 91.61 | 95.64 | 100.00 | 100.00 | 100.00 | 100.00 |
| 3 | 86.06 | 83.88 | 79.27 | 86.91 | 79.94 | 88.73 | 74.74 | 83.62 | 100.00 | 100.00 | 100.00 | 100.00 |
| 4 | 97.84 | 91.38 | 93.97 | 94.83 | 89.74 | 83.39 | 94.08 | 94.15 | 99.64 | 99.78 | 99.86 | 99.86 |
| 5 | 83.89 | 81.59 | 83.26 | 83.47 | 99.25 | 99.33 | 99.40 | 99.40 | 97.31 | 97.49 | 98.88 | 97.83 |
| 6 | 99.59 | 76.28 | 74.90 | 84.00 | 87.52 | 64.77 | 83.48 | 86.62 | 99.87 | 99.97 | 99.87 | 99.92 |
| 7 | 95.65 | 100.00 | 100.00 | 100.00 | 94.04 | 93.21 | 97.06 | 96.00 | 99.92 | 99.72 | 99.50 | 99.75 |
| 8 | 100.00 | 100.00 | 100.00 | 100.00 | 90.21 | 92.30 | 78.68 | 92.44 | 97.89 | 100.00 | 100.00 | 100.00 |
| 9 | 100.00 | 100.00 | 100.00 | 100.00 | 79.94 | 84.39 | 86.73 | 88.96 | 99.65 | 99.97 | 99.97 | 99.98 |
| 10 | 84.07 | 85.11 | 85.01 | 85.63 | 98.81 | 98.96 | 97.04 | 99.08 | ||||
| 11 | 82.16 | 90.78 | 72.45 | 91.35 | 96.14 | 95.39 | 100.00 | 98.12 | ||||
| 12 | 93.54 | 72.79 | 35.20 | 88.61 | 86.73 | 99.95 | 100.00 | 98.23 | ||||
| 13 | 99.50 | 99.50 | 99.50 | 99.50 | 98.24 | 97.04 | 97.15 | 97.69 | ||||
| 14 | 80.95 | 99.76 | 99.92 | 100.00 | 98.12 | 93.05 | 93.99 | 95.77 | ||||
| 15 | 76.64 | 79.53 | 75.59 | 77.17 | 95.75 | 99.53 | 99.90 | 99.93 | ||||
| 16 | 95.45 | 97.73 | 86.36 | 94.32 | 99.06 | 100.00 | 100.00 | 99.89 | ||||
| OA(%) | 85.37 | 85.50 | 80.59 | 87.98 | 88.92 | 84.77 | 88.08 | 91.75 | 98.07 | 99.44 | 99.54 | 99.57 |
| AA(%) | 90.64 | 89.03 | 85.48 | 90.97 | 87.79 | 85.81 | 87.80 | 91.18 | 97.95 | 98.80 | 99.13 | 99.13 |
| Kappa | 0.8340 | 0.8344 | 0.7802 | 0.8629 | 0.8539 | 0.7987 | 0.8433 | 0.8908 | 0.9785 | 0.9944 | 0.9948 | 0.9953 |
| T | Global | Single-scale | Multiscale | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCA | K PCA | S PCA | P-SKPCA | K-SKPCA | M-SKPCA | MS PCA | P-MSKPCA | K-MSKPCA | M-MSKPCA | 3-MSKPCA | ||
| Indian Pines | 5 | 46.39 | 44.61 | 77.21 | 77.78 | 73.78 | 75.19 | 78.98 | 81.38 | 79.82 | 80.68 | 83.56 |
| 10 | 55.65 | 56.85 | 86.49 | 86.54 | 84.86 | 83.82 | 87.18 | 88.03 | 86.90 | 87.69 | 89.15 | |
| 20 | 62.70 | 63.82 | 92.94 | 93.24 | 91.78 | 90.93 | 93.49 | 94.20 | 92.82 | 93.60 | 94.85 | |
| 30 | 66.27 | 65.54 | 95.06 | 95.16 | 93.69 | 94.27 | 95.18 | 95.81 | 95.29 | 95.71 | 96.33 | |
| Pavia Univ. | 5 | 65.26 | 60.04 | 75.43 | 78.62 | 77.02 | 75.67 | 84.29 | 84.90 | 83.17 | 86.99 | 87.95 |
| 10 | 70.00 | 65.27 | 85.81 | 88.27 | 86.54 | 89.21 | 91.61 | 92.05 | 91.81 | 92.84 | 93.76 | |
| 20 | 75.79 | 69.38 | 89.99 | 92.64 | 92.00 | 93.92 | 94.90 | 94.74 | 94.20 | 96.10 | 95.96 | |
| 30 | 76.13 | 69.63 | 91.50 | 93.92 | 93.26 | 95.05 | 95.75 | 96.10 | 95.27 | 97.01 | 97.13 | |
| Salinas | 5 | 81.87 | 79.00 | 94.69 | 94.87 | 99.28 | 98.38 | 96.87 | 96.86 | 99.33 | 98.87 | 99.38 |
| 10 | 85.25 | 83.39 | 97.16 | 97.33 | 99.35 | 99.03 | 97.85 | 98.16 | 99.40 | 99.35 | 99.41 | |
| 20 | 87.77 | 85.66 | 98.66 | 99.06 | 99.54 | 99.54 | 99.03 | 99.20 | 99.61 | 99.60 | 99.63 | |
| 30 | 89.24 | 87.14 | 99.22 | 99.32 | 99.61 | 99.59 | 99.34 | 99.41 | 99.63 | 99.64 | 99.71 | |
| Dataset | PCA | KPCA | SPCA | P-SKPCA | K-SKPCA | M-SKPCA |
|---|---|---|---|---|---|---|
| Indian Pines | 0.1528 | 0.3637 | 0.7874 | 1.6754 | 1.7500 | 1.7904 |
| Pavia University | 0.6907 | 1.7024 | 3.5299 | 4.6813 | 5.0714 | 4.9277 |
| Salinas | 0.7874 | 1.914 | 3.4234 | 5.3664 | 5.8822 | 5.9086 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Su, H.; Shen, J. Hyperspectral Dimensionality Reduction Based on Multiscale Superpixelwise Kernel Principal Component Analysis. Remote Sens. 2019, 11, 1219. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11101219
Zhang L, Su H, Shen J. Hyperspectral Dimensionality Reduction Based on Multiscale Superpixelwise Kernel Principal Component Analysis. Remote Sensing. 2019; 11(10):1219. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11101219
Chicago/Turabian StyleZhang, Lan, Hongjun Su, and Jingwei Shen. 2019. "Hyperspectral Dimensionality Reduction Based on Multiscale Superpixelwise Kernel Principal Component Analysis" Remote Sensing 11, no. 10: 1219. https://0-doi-org.brum.beds.ac.uk/10.3390/rs11101219