Distal Unfolding of Ferricytochrome c Induced by the F82K Mutation
Abstract
:1. Introduction
2. Results and Discussion
2.1. Ferrous F82K Cyt C
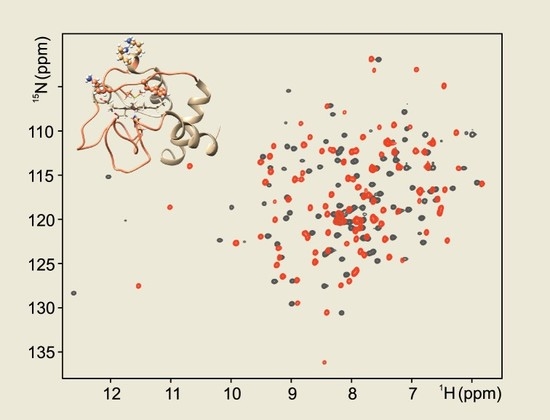
2.2. Ferric F82K Cyt C
2.3. F82A Cyt C
2.4. Additional Cyt C Variants
3. Materials and Methods
3.1. Site-Directed Mutagenesis
3.2. Protein Expression and Purification
3.3. NMR Sample Preparation
3.4. Magnetic Susceptibility Measurements
3.5. High-Resolution NMR Experiments
3.6. Assignment of F82K Cyt C
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| NMR | Nuclear Magnetic Resonance |
| Cyt c | Cytochrome c |
| EXSY | EXchange SpectroscopY |
| HSQC | Heteronuclear Single Quantum Coherence |
| CD | Circular Dichroism |
| MESNA | 2-Mercaptoethanesulfonic Acid |
| IPTG | IsoPropyl-β-D-ThioGalactopyranosid |
| OD | Optical Density |
References
- Scott, R.A.; Mauk, A.G. Cytochrome C: A Multidisciplinary Approach; University Science Books: Sausalito, CA, USA, 1996; p. 738. [Google Scholar]
- Banci, L.; Bertini, I.; Reddig, T.; Turano, P. Monitoring the conformational flexibility of cytochrome c at low ionic strength by 1H-NMR spectroscopy. Eur. J. Biochem. 1998, 256, 271–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banci, L.; Bertini, I.; Spyroulias, G.A.; Turano, P. The Conformational Flexibility of Oxidized Cytochrome c Studied through Its Interaction with NH3 and at High Temperatures. Eur. J. Inorg. Chem. 1998, 1998, 583–591. [Google Scholar] [CrossRef]
- Banci, L.; Berners-Price, S.J.; Bertini, I.; Clementi, V.; Luchinat, C.; Spyroulias, G.A.; Turano, P. Water-protein interaction in native and partially unfolded equine cytochrome c. Mol. Phys. 1998, 95, 797–808. [Google Scholar] [CrossRef]
- Bartalesi, I.; Bertini, I.; Ghosh, K.; Rosato, A.; Turano, P. The unfolding of oxidized c-type cytochromes: The instructive case of Bacillus pasteurii. J. Mol. Biol. 2002, 321, 693–701. [Google Scholar] [CrossRef]
- Berners-Price, S.J.; Bertini, I.; Gray, H.B.; Spyroulias, G.A.; Turano, P. The stability of the cytochrome c scaffold as revealed by NMR spectroscopy. J. Inorg. Biochem. 2004, 98, 814–823. [Google Scholar] [CrossRef]
- Hong, Y.; Muenzner, J.; Grimm, S.K.; Pletneva, E.V. Origin of the conformational heterogeneity of cardiolipin-bound cytochrome C. J. Am. Chem. Soc. 2012, 134, 18713–18723. [Google Scholar] [CrossRef] [Green Version]
- Muenzner, J.; Toffey, J.R.; Hong, Y.; Pletneva, E.V. Becoming a peroxidase: Cardiolipin-induced unfolding of cytochrome c. J. Phys. Chem. B 2013, 117, 12878–12886. [Google Scholar] [CrossRef] [Green Version]
- Assfalg, M.; Bertini, I.; Dolfi, A.; Turano, P.; Mauk, A.G.; Rosell, F.I.; Gray, H.B. Structural model for an alkaline form of ferricytochrome C. J. Am. Chem. Soc. 2003, 125, 2913–2922. [Google Scholar] [CrossRef]
- Baxter, S.M.; Fetrow, J.S. Hydrogen exchange behavior of [U-15N]-labeled oxidized and reduced iso-1-cytochrome c. Biochemistry 1999, 38, 4493–4503. [Google Scholar] [CrossRef]
- Fetrow, J.S.; Baxter, S.M. Assignment of 15N chemical shifts and 15N relaxation measurements for oxidized and reduced iso-1-cytochrome c. Biochemistry 1999, 38, 4480–4492. [Google Scholar] [CrossRef]
- Barker, P.D.; Bertini, I.; Del Conte, R.; Ferguson, S.J.; Hajieva, P.; Tomlinson, E.; Turano, P.; Viezzoli, M.S. A further clue to understanding the mobility of mitochondrial yeast cytochrome c: A (15)N T1rho investigation of the oxidized and reduced species. Eur. J. Biochem. 2001, 268, 4468–4476. [Google Scholar] [CrossRef] [PubMed]
- Assfalg, M.; Bertini, I.; Turano, P.; Mauk, A.G.; Winkler, J.R.; Gray, H.B. 15N-1H Residual dipolar coupling analysis of native and alkaline-K79A Saccharomyces cerevisiae cytochrome c. Biophys. J. 2003, 84, 3917–3923. [Google Scholar] [CrossRef] [Green Version]
- Sakamoto, K.; Kamiya, M.; Uchida, T.; Kawano, K.; Ishimori, K. Redox-controlled backbone dynamics of human cytochrome c revealed by 15N NMR relaxation measurements. Biochem. Biophys. Res. Commun. 2010, 398, 231–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, Y.; Weaver, M.L.; Hoke, K.R.; Pletneva, E.V. A Heme Propionate Staples the Structure of Cytochrome c for Methionine Ligation to the Heme Iron. Inorg. Chem. 2019, 58, 14085–14106. [Google Scholar] [CrossRef] [PubMed]
- Banci, L.; Bertini, I.; Bren, K.L.; Gray, H.B.; Turano, P. pH-dependent equilibria of yeast Met80Ala-iso-1-cytochrome c probed by NMR spectroscopy: A comparison with the wild-type protein. Chem. Biol. 1995, 2, 377–383. [Google Scholar] [CrossRef] [Green Version]
- Banci, L.; Bertini, I.; Rosato, A.; Varani, G. Mitochondrial cytochromes c: A comparative analysis. J. Biol. Inorg. Chem. 1999, 4, 824–837. [Google Scholar] [CrossRef]
- Wegerich, F.; Turano, P.; Allegrozzi, M.; Mohwald, H.; Lisdat, F. Cytochrome C mutants for superoxide biosensors. Anal. Chem. 2009, 81, 2976–2984. [Google Scholar] [CrossRef]
- Louie, G.V.; Pielak, G.J.; Smith, M.; Brayer, G.D. Role of phenylalanine-82 in yeast iso-1-cytochrome c and remote conformational changes induced by a serine residue at this position. Biochemistry 1988, 27, 7870–7876. [Google Scholar] [CrossRef]
- Bertini, I.; Luchinat, C.; Turano, P.; Battaini, G.; Casella, L. The magnetic properties of myoglobin as studied by NMR spectroscopy. Chemistry 2003, 9, 2316–2322. [Google Scholar] [CrossRef]
- Santucci, R.; Ascoli, F. The Soret circular dichroism spectrum as a probe for the heme Fe(III)-Met(80) axial bond in horse cytochrome c. J. Inorg. Biochem. 1997, 68, 211–214. [Google Scholar] [CrossRef]
- Rosell, F.I.; Ferrer, J.C.; Mauk, A.G. Proton-Linked Protein Conformational Switching: Definition of the Alkaline Conformational Transition of Yeast Iso-1-ferricytochrome c. JACS 1998, 120, 11234–11245. [Google Scholar] [CrossRef]
- Pollock, W.B.; Rosell, F.I.; Twitchett, M.B.; Dumont, M.E.; Mauk, A.G. Bacterial expression of a mitochondrial cytochrome c. Trimethylation of lys72 in yeast iso-1-cytochrome c and the alkaline conformational transition. Biochemistry 1998, 37, 6124–6131. [Google Scholar] [CrossRef] [PubMed]
- Turano, P. Insights into Partially Folded or Unfolded States of Metalloproteins from Nuclear Magnetic Resonance. Inorg. Chem. 2004, 43, 7945–7952. [Google Scholar] [CrossRef]
- Cacciatore, S.; Piccioli, M.; Turano, P. Electron self-exchange of cytochrome c measured via13C detected protonless NMR. J. Porphyr. Phthalocyanines 2013, 17, 142–149. [Google Scholar] [CrossRef]
- Bertini, I.; Turano, P.; Vila, A.J. Nuclear magnetic resonance of paramagnetic metalloproteins. Chem. Rev. 1993, 93, 2833–2932. [Google Scholar] [CrossRef]
- Banci, L.; Bertini, I.; Gray, H.B.; Luchinat, C.; Reddig, T.; Rosato, A.; Turano, P. Solution structure of oxidized horse heart cytochrome c. Biochemistry 1997, 36, 9867–9877. [Google Scholar] [CrossRef] [Green Version]
- Banci, L.; Bertini, I.; Bren, K.L.; Gray, H.B.; Sompornpisut, P.; Turano, P. Solution structure of oxidized Saccharomyces cerevisiae iso-1-cytochrome c. Biochemistry 1997, 36, 8992–9001. [Google Scholar] [CrossRef] [Green Version]
- Wuthrich, K. Protein structure determination in solution by NMR spectroscopy. J. Biol. Chem. 1990, 265, 22059–22062. [Google Scholar]
- Wider, G.; Wuthrich, K. NMR spectroscopy of large molecules and multimolecular assemblies in solution. Curr. Opin. Struct. Biol. 1999, 9, 594–601. [Google Scholar] [CrossRef]
- Kay, L.E.; Ikura, M.; Tschudin, R.; Bax, A. Three-dimensional triple-resonance NMR Spectroscopy of isotopically enriched proteins. J. Magn. Reson. 2011, 213, 423–441. [Google Scholar] [CrossRef]
- Döpner, S.; Hildebrandt, P.; Rosell, F.I.; Mauk, A.G. Alkaline Conformational Transitions of Ferricytochrome c Studied by Resonance Raman Spectroscopy. JACS 1998, 120, 11246–11255. [Google Scholar] [CrossRef]
- Hong, X.L.; Dixon, D.W. NMR study of the alkaline isomerization of ferricytochrome c. FEBS Lett. 1989, 246, 105–108. [Google Scholar] [CrossRef] [Green Version]
- Ferrer, J.C.; Guillemette, J.G.; Bogumil, R.; Inglis, S.C.; Smith, M.; Mauk, A.G. Identification of Lys79 as an iron ligand in one form of alkaline yeast iso-1-ferricytochrome c. JACS 1993, 115, 7507–7508. [Google Scholar] [CrossRef]
- Jeng, W.Y.; Chen, C.Y.; Chang, H.C.; Chuang, W.J. Expression and characterization of recombinant human cytochrome c in E. coli. J. Bioenerg. Biomembr. 2002, 34, 423–431. [Google Scholar] [CrossRef]
- Rivera, M.; Walker, F.A. Biosynthetic preparation of isotopically labeled heme. Anal. Biochem. 1995, 230, 295–302. [Google Scholar] [CrossRef]
- Turano, P.; Lalli, D.; Felli, I.C.; Theil, E.C.; Bertini, I. NMR reveals pathway for ferric mineral precursors to the central cavity of ferritin. Proc. Natl. Acad. Sci. USA 2010, 107, 545–550. [Google Scholar] [CrossRef] [Green Version]
- Winkler, J.R.; Wittung-Stafshede, P.; Leckner, J.; Malmstrom, B.G.; Gray, H.B. Effects of folding on metalloprotein active sites. Proc. Natl. Acad. Sci. USA 1997, 94, 4246–4249. [Google Scholar] [CrossRef] [Green Version]
- Banci, L.; Bertini, I.; Huber, J.G.; Spyroulias, G.A.; Turano, P. Solution structure of reduced horse heart cytochrome c. J. Biol. Inorg. Chem. 1999, 4, 21–31. [Google Scholar] [CrossRef]
- Bertini, I.; Chevance, S.; Del Conte, R.; Lalli, D.; Turano, P. The anti-apoptotic Bcl-x(L) protein, a new piece in the puzzle of cytochrome c interactome. PLoS ONE 2011, 6, e18329. [Google Scholar] [CrossRef]
- Liptak, M.D.; Fagerlund, R.D.; Ledgerwood, E.C.; Wilbanks, S.M.; Bren, K.L. The proapoptotic G41S mutation to human cytochrome c alters the heme electronic structure and increases the electron self-exchange rate. J. Am. Chem. Soc. 2011, 133, 1153–1155. [Google Scholar] [CrossRef] [Green Version]
- Josephs, T.M.; Liptak, M.D.; Hughes, G.; Lo, A.; Smith, R.M.; Wilbanks, S.M.; Bren, K.L.; Ledgerwood, E.C. Conformational change and human cytochrome c function: Mutation of residue 41 modulates caspase activation and destabilizes Met-80 coordination. J. Biol. Inorg. Chem. 2013, 18, 289–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karsisiotis, A.I.; Deacon, O.M.; Wilson, M.T.; Macdonald, C.; Blumenschein, T.M.; Moore, G.R.; Worrall, J.A. Increased dynamics in the 40–57 Omega-loop of the G41S variant of human cytochrome c promote its pro-apoptotic conformation. Sci. Rep. 2016, 6, 30447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno-Beltran, B.; Guerra-Castellano, A.; Diaz-Quintana, A.; Del Conte, R.; Garcia-Maurino, S.M.; Diaz-Moreno, S.; Gonzalez-Arzola, K.; Santos-Ocana, C.; Velazquez-Campoy, A.; De la Rosa, M.A.; et al. Structural basis of mitochondrial dysfunction in response to cytochrome c phosphorylation at tyrosine 48. Proc. Natl. Acad. Sci. USA 2017, 114, E3041–E3050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciambellotti, S.; Turano, P. Structural Biology of Iron-Binding Proteins by NMR Spectroscopy. Eur. J. Inorg. Chem. 2019, 2019, 569–576. [Google Scholar] [CrossRef]
- Bertini, I.; Cavallaro, G.; Rosato, A. A structural model for the adduct between cytochrome c and cytochrome c oxidase. J. Biol. Inorg. Chem. 2005, 10, 613–624. [Google Scholar] [CrossRef]
- Bertini, I.; Cavallaro, G.; Rosato, A. Principles and patterns in the interaction between mono-heme cytochrome c and its partners in electron transfer processes. Metallomics 2011, 3, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Lo, T.P.; Komar-Panicucci, S.; Sherman, F.; McLendon, G.; Brayer, G.D. Structural and functional effects of multiple mutations at distal sites in cytochrome c. Biochemistry 1995, 34, 5259–5268. [Google Scholar] [CrossRef]
- Rosell, F.I.; Harris, T.R.; Hildebrand, D.P.; Dopner, S.; Hildebrandt, P.; Mauk, A.G. Characterization of an alkaline transition intermediate stabilized in the Phe82Trp variant of yeast iso-1-cytochrome c. Biochemistry 2000, 39, 9047–9054. [Google Scholar] [CrossRef]
- Banci, L.; Bertini, I.; Luchinat, C.; Turano, P. Biological Inorganic Chemistry: Structure and Reactivity, 1st ed.; University Science Books: Sausalito, CA, USA, 2006; p. 739. [Google Scholar]
- Sinibaldi, F.; Howes, B.D.; Droghetti, E.; Polticelli, F.; Piro, M.C.; Di Pierro, D.; Fiorucci, L.; Coletta, M.; Smulevich, G.; Santucci, R. Role of lysines in cytochrome c-cardiolipin interaction. Biochemistry 2013, 52, 4578–4588. [Google Scholar] [CrossRef] [Green Version]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lalli, D.; Rosa, C.; Allegrozzi, M.; Turano, P. Distal Unfolding of Ferricytochrome c Induced by the F82K Mutation. Int. J. Mol. Sci. 2020, 21, 2134. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21062134
Lalli D, Rosa C, Allegrozzi M, Turano P. Distal Unfolding of Ferricytochrome c Induced by the F82K Mutation. International Journal of Molecular Sciences. 2020; 21(6):2134. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21062134
Chicago/Turabian StyleLalli, Daniela, Camilla Rosa, Marco Allegrozzi, and Paola Turano. 2020. "Distal Unfolding of Ferricytochrome c Induced by the F82K Mutation" International Journal of Molecular Sciences 21, no. 6: 2134. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21062134