1. Introduction
Diabetes is one of the most common human diseases and has become a significant public health concern worldwide. There were approximately 450 million people diagnosed with diabetes that resulted in around 1.37 million deaths globally in 2017 [
1]. More than 100 million US adults live with diabetes, and diabetes was the seventh leading cause of death in the US in 2020 [
2]. One in ten US adults have diabetes now, and if the current trend continues, it is projected that as many as one in three US adults could have diabetes by 2050 [
2]. Diabetes patients are at elevated risk of developing health complications such as kidney failure, vision loss, heart disease, stroke, premature death, and amputation of feet or legs, which can lead to dysfunction and chronic damage of tissue [
3]. In addition, there are substantial economic costs associated with the disease. The total estimated price of diagnosed diabetes in the US increased to USD 237 billion in 2017 from USD 188 billion in 2012. The excess medical costs per person associated with diabetes increased to USD 9601 from USD 8417 during the same period [
2]. Additionally, there could be productivity loss due to diabetic patients in the workforce.
An individual at high risk of diabetes may not be aware of the risk factors associated with it. Given the high prevalence and severity of diabetes, researchers are interested in finding the most common risk factors of diabetes, as it could be due to a combination of several reasons. Determining the risk factors and early prediction of diabetes have been vital in reducing diabetes complications [
4,
5] and economic burden [
6] and is beneficial from both clinical practice and public health perspectives [
7]. Similarly, studies find that screening high-risk individuals identifies the population groups in which implementing measures aimed at preventing diabetes will be the most beneficial [
8]. Early intervention may help prevent complications and improve quality of life and are essential in designing effective prevention strategies [
9,
10]. There is growing evidence that lifestyle modification prevents or delays type 2 diabetes [
11]. The main risk factors of diabetes are considered to be an unhealthy diet, aging, family history, ethnic groups, obesity, sedentary lifestyle, and previous history of gestational diabetes [
6,
7,
12]. Previous studies have also reported that sex, body mass index (BMI), pregnancy, and metabolic status are associated with diabetes [
13,
14].
Prediction models can screen pre-diabetes or people with an increased risk of developing diabetes to help decide the best clinical management for patients. Numerous predictive equations have been suggested to model the risk factors of incident diabetes [
15,
16,
17]. For instance, Heikes et al. [
18] studied a tool to predict the risk of diabetes in the US using undiagnosed and pre-diabetes data, and Razavian et al. [
19] developed logistic regression-based prediction models for type 2 diabetes occurrence. These models also help screen individuals to posit individuals who are at a high risk of having diabetes. Zou et al. [
20] used machine learning methods to predict diabetes in Luzhou, China, and a five-fold cross-validation was used to validate the models. Nguyen et al. [
5] predict the onset of diabetes employing deep learning algorithms suggesting that sophisticated methods may improve the performance of models. In contrast, several other studies have shown that logistic regression performs as least as well as machine learning techniques for disease risk prediction ([
21,
22], for example). Similarly, Anderson et al. [
23] used logistic regression along with machine learning algorithms and found a higher accuracy with the logistic regression model. These are mainly based on assessing risk factors of diabetes, such as household and individual characteristics; however, the lack of an objective and unbiased evaluation is still an issue [
24]. Additionally, there is growing concern that those predictive models are poorly developed due to inappropriate selection of covariates, missing data, small samples size, and wrongly specified statistical models [
25,
26]. To this end, only a few risk prediction models have been routinely used in clinical practice. The reliability and quality of these predictive tools and equations show significant variation depending on geography, available data, and ethnicity [
5]. Risk factors for one ethnic group may not be generalized to others; for example, the prevalence of diabetes is reported to be higher among the Pima Indian community. Therefore, this study uses the Pima Indian dataset to predict if an individual is at risk of developing diabetes based on specific diagnostic factors [
27,
28].
We consider a combined approach of logistic regression and machine learning to predict the risk factors of type 2 diabetes mellitus. The logistic regression compares several prediction models for predicting diabetes. We used various selection criteria popular in the literature such as
AIC,
BIC, Mallows’ Cp, adjusted
, and forward and backward selection to identify the significant predictors. We then exploit the classification tree, a widely used machine learning technique with considerable classification power [
5,
20,
25], to predict diabetes incidence in several previous studies. Our paper also contributes to the broad literature on risk factors of diabetes. We extend previous research by exploring traditional econometric models and simple machine learning algorithms to build on the literature on diabetes prediction. Our analysis finds five main predictors of diabetes: glucose, pregnancy, body mass index, age, and diabetes pedigree function. These risk factors of diabetes identified by the logistic regression were validated by the decision tree and could help classify high-risk individuals and prevent, diagnose and manage diabetes. A regression model containing the above five predictors yields a prediction accuracy of 77.73% and a cross-validation error rate of 22.65%. This study helps inform policymakers and design the health policy to curb the prevalence of diabetes. The methods exhibiting the best classification performance vary depending on the data structure; therefore, future studies should be cautious using a single model or approach for diabetes risk prediction.
The remainder of this article proceeds as follows. The next section discusses data and summary statistics. We then describe the methods. This is followed by a presentation of the results. The last section concludes our study.
2. Data and Summary Statistics
Several factors might be related to diabetes, including blood pressure, pregnancies for women, age and body mass index, etc. As a component of diabetes management, it would be helpful to know which variables are related to diabetes. We use the Pima Indian dataset, made available by the National Institute of Diabetes at the Johns Hopkins University, as a test case to predict the risk factors associated with diabetes. The Pima are American Indians that live along the Gila River and Salt River in Southern Arizona. Each observation in the dataset represents an individual patient and includes information on the patient’s diabetes classification, along with the various medical attributes such as the number of pregnancies, plasma glucose concentration, tricep skinfold thickness, body mass index (BMI), diastolic blood pressure, 2-hour serum insulin (serum-insulin), age and diabetes pedigree function. Our response variable, diabetes, would take the value of 1 if an individual were diagnosed with type 2 diabetes and 0 otherwise. There are 268 (34.9%) diabetes patients in our sample. Five covariates, insulin, glucose, BMI, skin thickness, and blood pressure, contain at least one missing value (indexed by zero), which is not meaningful. So, we replaced all those zeros with the corresponding median values. We analyzed data using statistical program R version 4.0.5 [
29].
Table 1 presents descriptive statistics for all predictors after median value imputation for missing values.
Table 1 shows that BP, BMI, and skin-thickness have almost the same mean and median. The variable pedigree has the lowest standard deviation while insulin has the highest standard deviation, implying that pedigree has the least variability and insulin has the highest variability present in their distributions. Our objective is to identify a subset of covariates most suitable for inclusion in a predictive model for diabetes. Including only a few predictors can lead to omitted variable bias, and too many predictors can reduce the precision. There are numerous methods available for developing appropriate predictive equations. Among them, we implement logistic regression and a classification tree—a machine learning technique.
4. Results
Figure 1 is the correlation plot, which gives the strength of the correlations between different pairs of the predictors. The pairwise correlations (
r) between pregnancy and age (0.59) and between BMI and skin thickness (0.54) are (
) high compared to other pairs, indicating these two pairs of predictors are significantly correlated.
4.1. Estimates of Logistic Regression Models
We begin by looking at the predictors of the prevalence of diabetes using the logistic regression model. We tried all possible combinations of predictors and then compared each model using the goodness of fit test and model selection criteria. We outlined five candidate models while performing the goodness of fit test.
Table 2 shows results from fitting the logistic regression models, in which we compared results across five different candidate regression specifications and a null model. Model 1 represents estimates from the null model, while model 2 estimates the full model. Since the residual deviance is decreased significantly with the inclusion of predictor variables, we can say that the model with the inclusion of predictor variables is better than the null model. Based on the full model, the variables skin thickness, blood pressure, and insulin are not significant predictors of diabetes at 5% significance level. Model 3 is the estimate of the logistic regression, omitting the non-significant variable from model 2 as we want to ensure a better estimation of the regression coefficients. Model 4, model 5, and model 6 add different interaction terms as we observe interacting age with pregnancy, glucose, and insulin, and insulin with pedigree provides more meaningful results.
We proceed to obtain the best possible model specified by the best subset selection method. For this purpose, we used AIC, BIC, log-likelihood, , adjusted , and criteria. Model 6 has the smallest AIC and the largest and log-likelihood values, suggesting a better fit; however, model 4 has the smallest BIC, indicating ambiguity. Notice that and log-likelihood values increase continuously with each additional variable included in the model. These two criteria are not much helpful for selecting the best fit model since these statistics do not indicate which and how many predictor variables to include. Additionally, we have to be cautious about the possibility of obtaining an over-fitted model while including several covariates and interaction terms that could lead to a problem with near co-linearity.
We drew interaction plots to better understand the nature of the interactions between age and glucose and between age and pregnancy.
Figure 2 and
Figure 3 show how the variation in one of the predictor variable changes the response given a fixed value of the interacting variable.
As seen in
Figure 2, the probability of diabetes is higher for high-age groups when the glucose level is below 160 mg/dL (approx). However, when the glucose level is above 160 mg/dL, the probability of diabetes is higher for low-age groups and lower for high-age group women. The second plot in the
Figure 2 shows that the probability of diabetes is the highest for women with high glucose levels (185 mg/dL). The lower the glucose level, the lower the probability of diabetes.
Figure 3 shows the interaction between age and pregnancy. As seen in the plot, there is an inverse relationship between age and pregnancy in the prediction of diabetes. When the number of pregnancies is greater than 7, the probability of diabetes is higher for low age groups (<55 years) and less for higher age group women. This relationship is the opposite when the number of pregnancies for a woman is less than 7. On the other hand, if we keep pregnancy fixed and vary age, the probability of diabetes is higher for the higher age group (>55 years) who have no children and the lowest for the women of age
years who have had six pregnancies. At the same time, the probability of diabetes incidence remains lower if a woman is in the low-age group with fewer than six pregnancies.
The plots of residual sum of square (RSS), adjusted
,
, and
BIC for all models together will help us decide the best fit model to select.
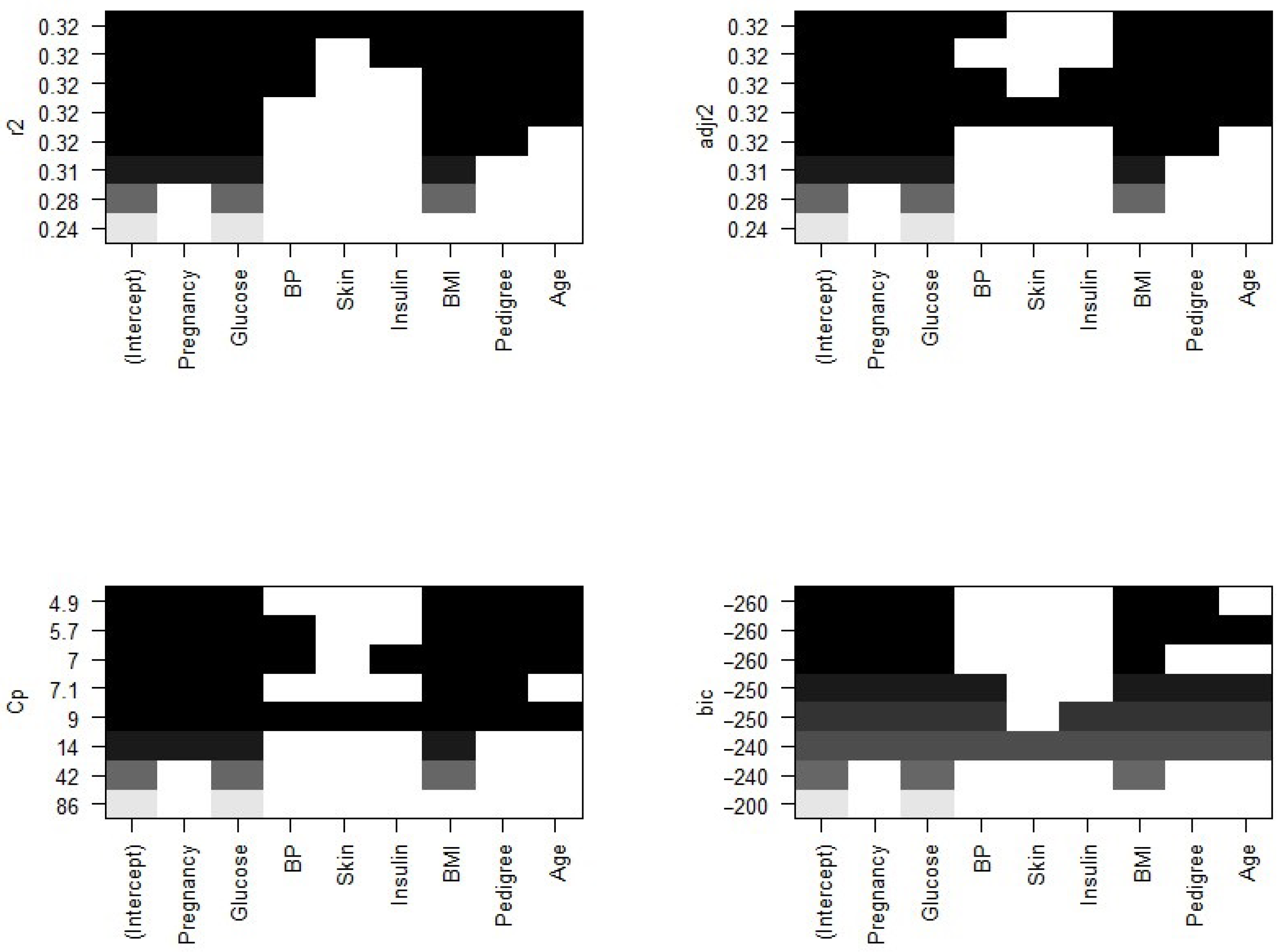
Figure 4 plots the relationship between selection criteria versus a number of predictors in models. We observed that different criteria suggest different sized models that fit best. According to RSS criteria, five or six variable models fit the data better. Adjusted
selected a six-variable model, while
BIC and
suggest four- and five-variable models, respectively.
Now, we need to know which of the predictors should be selected. According to the
BIC criteria, there are only four predictors: pregnancy, glucose, BMI, and pedigree. The model with these four predictors has the minimum
BIC value (results not shown). According to Mallows’ Cp criteria, we have five significant predictor variables: pregnancy, glucose, BMI, pedigree, and age. If we look at the adjusted
plot, six variables, pregnancy, glucose, BP, BMI, pedigree, and age, are significant. The coefficients associated with the four predictors given by
BIC and that for five predictors associated with the Mallows’ Cp criterion are given in
Figure 5.
We further explored models suggested by the forward and backward stepwise selection method. For this, we used the regsubsets() function in R. The forward and backward method both agree on the five predictor variables, pregnancy, glucose, BMI, pedigree, and age, which is consistent with the other selection criteria to give the best fit model.
4.2. Classification Tree and Prediction Accuracy
Our variable outcome,
diabetes, is a binary numeric variable indexed by 1 and 0. We changed this variable into a qualitative variable to obtain a classification tree. Accordingly, we recoded variable labels so that “Yes” indicates the diabetes patient and “No” otherwise.
Figure 5 shows the classification tree with thirteen terminal nodes and seven predictors. Deviance in the classification tree, as in James et al. [
30], is given by
where
is the number of sample points in the
terminal node that belong to the
kth class. Small deviance indicates a good fit for the (training) data. The residual mean deviance measures the goodness of fit and is simply the deviance divided by
.
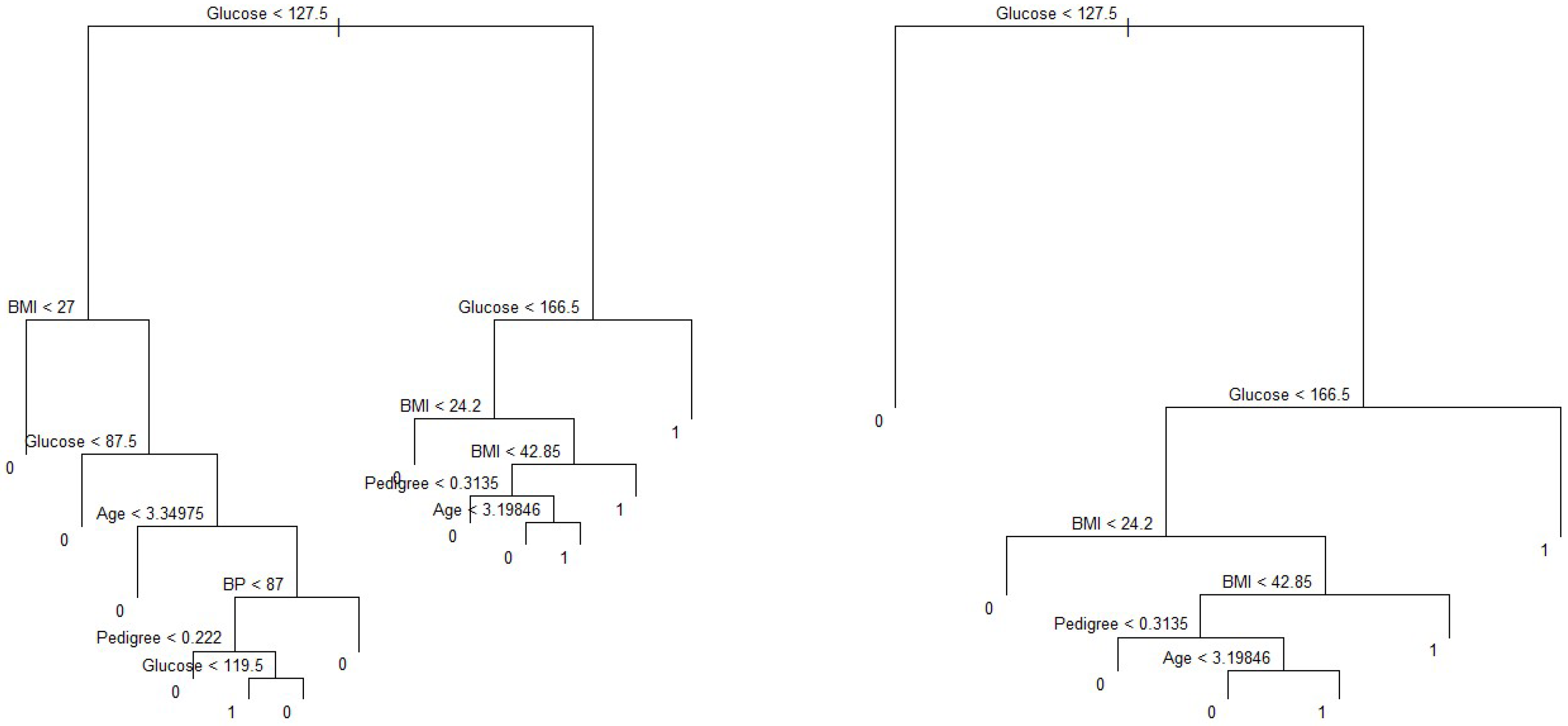
Figure 5 shows a classification tree using all predictors available in the dataset. Glucose is the root node, suggesting that glucose is an important predictor of diabetes while BMI and age are also key variables in this method. Our results from this method are also in line with Wu et al. (2021), who indicated that glucose, BMI, and age were the top three predictors of diabetes. Residual mean deviance corresponding to
Figure 6 is 601/755 = 0.796.
We consider the full-sized tree with thirteen terminal nodes to check the performance of different trees and subtrees. In order to evaluate the performance of a classification tree, we need to estimate the test error rate. For this, we split the dataset into a training set and a test set. The training dataset contains
of the observations and the test set contains the remaining
observations, which were assigned randomly. We built trees using the training set and evaluated their performance on the test data using the
predict() function in statistical program R. The confusion matrix corresponding to the thirteen-node tree is given in
Table 3.
The prediction accuracy rate of this thirteen-node tree is , meaning that this tree cannot predict accurately of the time. We can check the performance of its subtrees by pruning for better predictive ability.
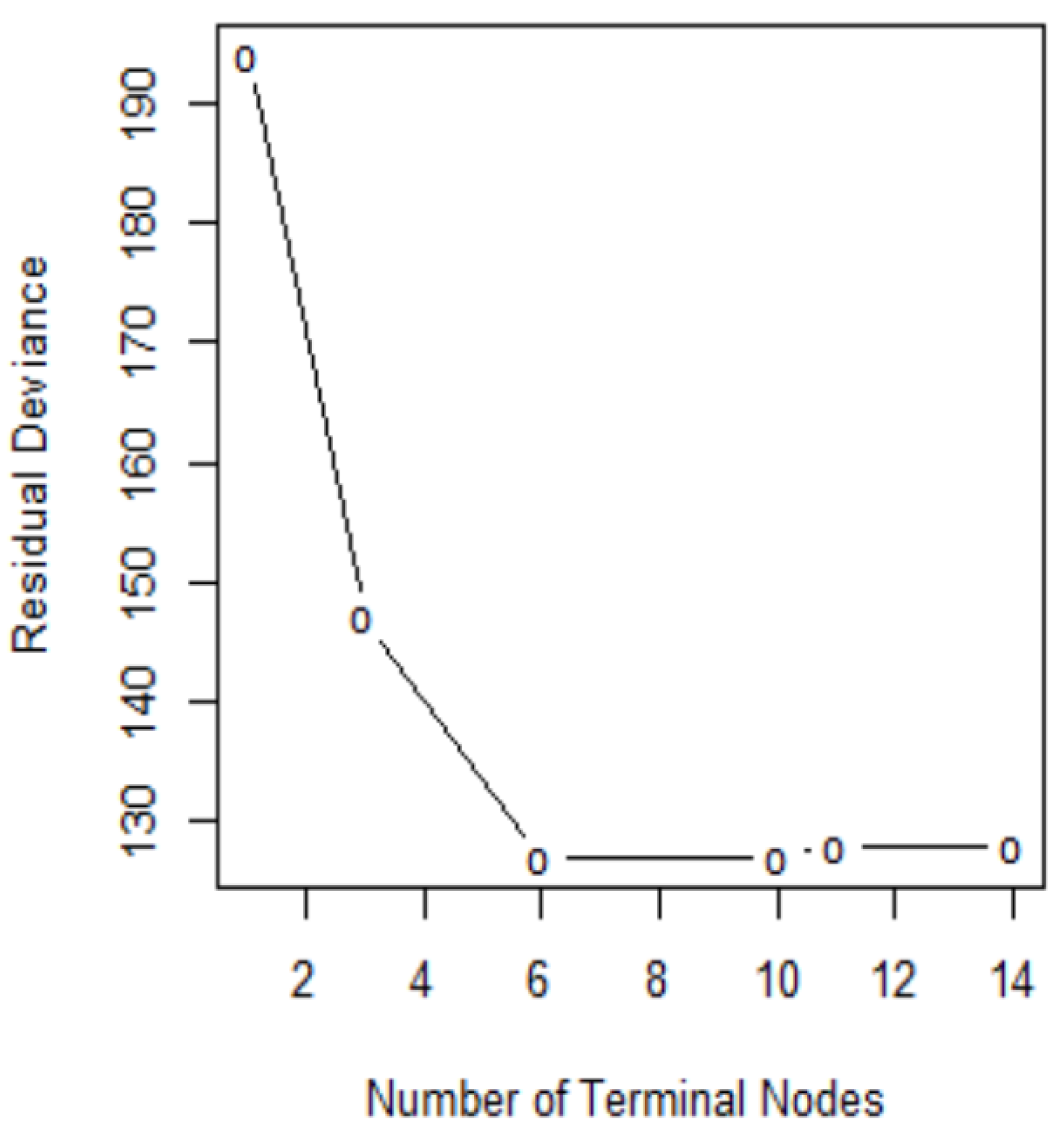
In order to determine the optimal level of tree complexity, we employed the function
cv.tree() in R. Cost complexity pruning was also used to select a sequence of trees for consideration. We applied the
prune.misclass() function in R to prune the tree. We check for the performance of the trees with 14, 10, and 6 terminal nodes as suggested by the
cv.tree()function.
Figure 7 shows pruning plots which were determined by the size (the number of terminal nodes of tree) and
. The vertical axis represents residual deviance. The model with the lowest residual deviance is preferred, as shown in
Table 4.
We observe that subtrees with ten and six terminal nodes have the lowest residual deviance; however, a tree with ten nodes may cause over-fitting. Thus, a six-node tree could be a better choice. The residual deviance for this tree is 127, and the optimal value of the tuning parameter is 3.25. The prediction accuracy corresponding to this six-terminal node tree is
(
Table 5). It means
of the test observations can be correctly classified with only four predictors: glucose, BMI, pedigree, and age. It also shows the improved accuracy compared to that of full-sized tree.
On the other hand, the predictors associated with the ten-node subtree are also the same—that is, glucose, BMI, pedigree and age, (
Figure 8), with a prediction accuracy of
, which is same as that obtained from a six-node subtree.
4.3. Proposed Diabetes Predictive Equation for Pima Indians
For the sake of completeness, we also calculated the prediction accuracy and cross-validation errors of the models proposed based on the logistic regression (
Table 6). Model 6 has the highest prediction accuracy of
among all models considered in this study, and it has a classification error rate equal to
, which is better than that of the other candidate models.
To check the model’s predictive ability, we ran eight-fold cross-validation for the model, which gives a prediction error rate equal to
. This is slightly higher than that given in
Table 7. Results provide evidence that model 6 with four interaction terms performs better than any combinations of covariates for the Pima Indian diabetes dataset. Thus, we propose the following model:
5. Discussion
Diabetes has become one of the leading causes of human death in recent decades. The incidence of diabetes has been continuously increasing every year due to several reasons including eating habits, sedentary lifestyle, and prevalence of unhealthful foods. Diabetes prediction model can contribute to the decision-making process in clinical management. Knowing the potential risk factors and identifying individuals at high risk in the early stages may aid in diabetes prevention. A host of prediction models for diabetes have been developed and applied, out of which logistic regression [
21] and a machine learning algorithm-based classification tree [
20] are among the most popular methods. Habibi et al. [
6] suggest that a simple machine learning algorithm, a classification tree, could be used to screen diabetes without using a laboratory. However, the validity of these models for different locations, populations with different diets, lifestyle, races, and genetic makeup is still unknown. Additionally, only a limited number of the reliable predictive equation has been suggested for Pima Indian Women. Their prediction performance and validity vary considerably. To fill this gap in the literature, this study used logistic regression and a classification tree from the Pima Indian dataset to identify the important factors for type 2 diabetes. We selected variables based on the goodness of fit test and model selection criteria such as
AIC,
BIC, and Mallows’ Cp. The decision trees were plotted, and the prediction accuracy and cross-validation error rate were calculated for the purpose of validation. Variables selected from the logistic regression and decision tree are very similar, suggesting that the variable identified helps predict diabetes and may be used as a decision tool.
A higher BMI, reduced insulin secretion and action, a family history of diabetes, blood pressure, smoking status, and pregnancy status are considered common risk factors for type 2 diabetes [
33,
34]. Our study shows five main predictors for diabetes are frequency of pregnancies in women, glucose, pedigree, BMI, and age. These variables were also used in previous studies to predict diabetes. For example, Bays et al. [
35] reported that increased BMI was associated with an increased risk of diabetes mellitus. A study in Finland developed a diabetes risk score to predict diabetes and found that age, parental history of diabetes, BMI, high blood sugar level, and physical activities are among the predictors of diabetes [
36]. People with higher glucose are more likely to develop diabetes. It may be because glucose is associated with insulin response [
37]. Lyssenko et al. [
33] reported that a family history of diabetes could double the risk of the disease. The pedigree provides a synthesis of the diabetes mellitus history in ancestors and the genetic relationship with the subject. It utilizes information from a person’s family history to predict how likely a subject can get diabetes. A higher BMI results in obesity, which could increase the fat content of the pancreas and might affect the function of pancreatic cells. Obesity could also lead to insulin resistance [
38,
39]. Age is a risk factor for the onset of diabetes. Pancreatic cells lead to the decline of glucose sensitivity and impaired insulin secretion with aging [
40]. The validation shows that our model has a relatively good predictive performance. The prediction accuracy of 78% from our preferred specification is close to previous studies on diabetes risk factors. For example, Lyssenko et al. [
33] report accuracy rates of 74% to 77% for two different locations in the study of diabetes risk factors. Zou et al. [
20] predict diabetes with accuracy values of 77% and 81% for the Pima Indian and Luzhou datasets, respectively. As indicated by Wilson et al. [
41], we also found that complex models are not necessary to predict diseases; instead, logistic regression and classification tree techniques can be equally useful in predicting diabetes. However, validation of the proposed model among different groups of the population should be carried out.
We acknowledge the limitations of our analysis. First, only a few predictors were considered to predict the risk of diabetes due to data limitations. Thus, our conclusion may not be generalizable to larger datasets with several predictors. Second, even the best predictive models and variable selection processes may yield different results according to location, type of dataset, and the algorithms used. Finally, we replaced missing values with medians of the respective variables which, although a common practice, could alter results. Future studies could incorporate several other risk factors such as genetic traits, gender, socio-economic status, physical activities, smoking, health information and attitude, food consumption, and spending to predicting diabetes in a more generalized population.