In Silico Prediction and Prioritization of Novel Selective Antimicrobial Drug Targets in Escherichia coli
Abstract
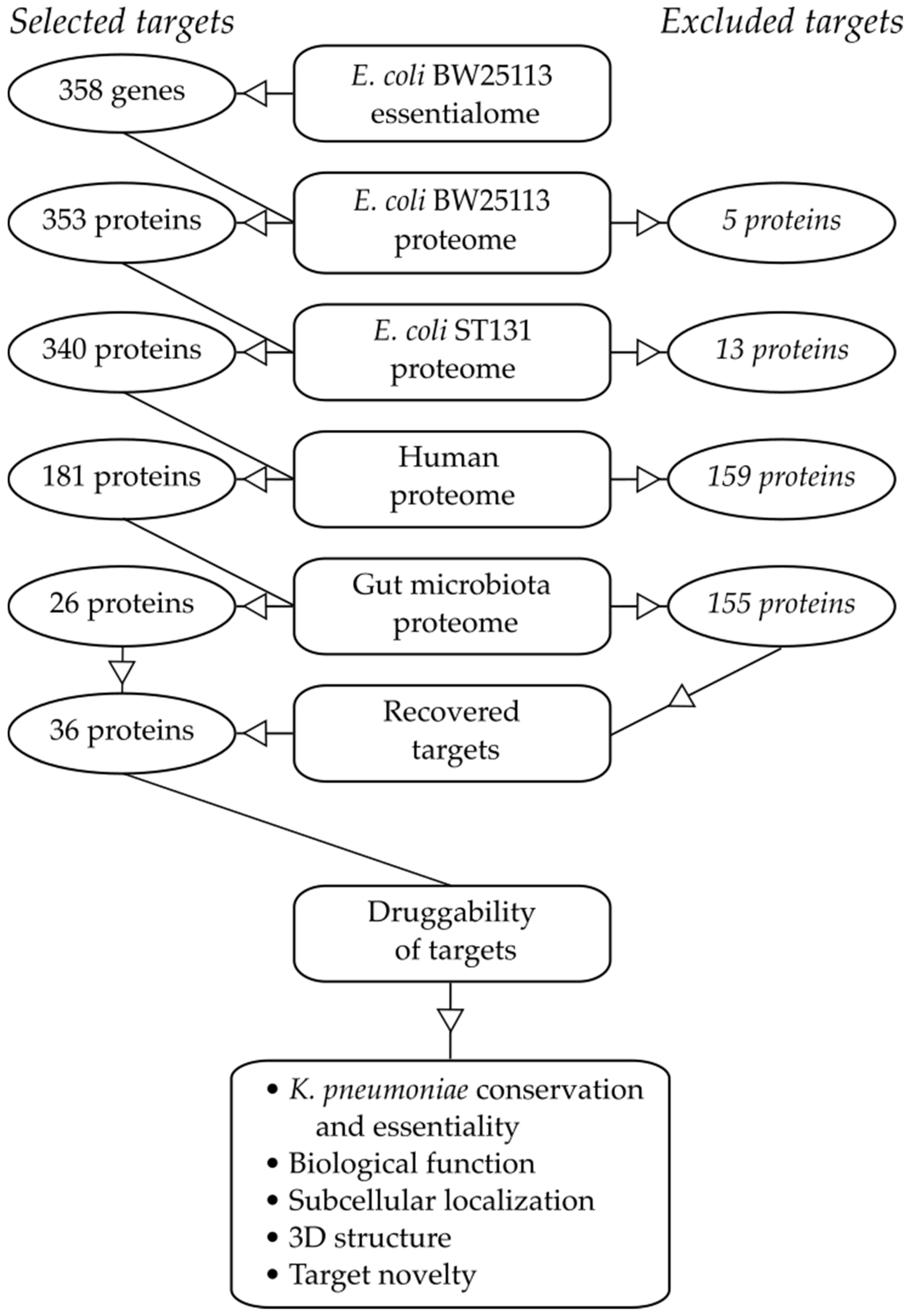
:1. Introduction
2. Results
2.1. Essential Genes in the Target Pathogen
2.2. Similarity to Proteins in Mammalian Hosts
2.3. Similarity to Proteins in Beneficial Taxa of the Gut Microbiota
2.4. Target Essentiality and Conservation in K. pneumoniae
2.5. Biological Function of Selected Targets
2.6. Target Localization
2.7. Existence of Known Inhibitors
2.8. Target Structure
3. Discussion
4. Materials and Methods
4.1. Protein Essentiality in E. coli
4.2. Protein Homology in E. coli ST131, Humans and Gut Beneficial Taxa
4.3. Protein Conservation and Essentiality in K. pneumoniae
4.4. Identification of Inhibitors by Screening Scientific Literature
4.5. Protein SCL and 3D Structure
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lewis, K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 2013, 12, 371–387. [Google Scholar] [CrossRef] [PubMed]
- Provenzani, A.; Hospodar, A.R.; Meyer, A.L.; Leonardi Vinci, D.; Hwang, E.Y.; Butrus, C.M.; Polidori, P. Multidrug-resistant Gram-negative organisms: A review of recently approved antibiotics and novel pipeline agents. Int. J. Clin. Pharm. 2020, 42, 1016–1025. [Google Scholar] [CrossRef] [PubMed]
- Martin-Loeches, I.; Dale, G.E.; Torres, A. Murepavadin: A new antibiotic class in the pipeline. Expert Rev. Anti. Infect. Ther. 2018, 16, 259–268. [Google Scholar] [CrossRef]
- World Health Organization. World Health Organization Releases Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Available online: https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf. (accessed on 24 May 2021).
- Köhler, C.D.; Dobrindt, U. What defines extraintestinal pathogenic Escherichia coli? Int. J. Med. Microbiol. 2011, 301, 642–647. [Google Scholar] [CrossRef]
- Pitout, J.D.D.; DeVinney, R. Escherichia coli ST131: A multidrug-resistant clone primed for global domination. F1000Research 2017, 6, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Buffie, C.G.; Pamer, E.G. Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 2013, 13, 790–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez, C.; Taminiau, B.; Van Broeck, J.; Delmée, M.; Daube, G. Clostridium difficile infection and intestinal microbiota interactions. Microb. Pathog. 2015, 89, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Hills, R.D.; Pontefract, B.A.; Mishcon, H.R.; Black, C.A.; Sutton, S.C.; Theberge, C.R. Gut microbiome: Profound implications for diet and disease. Nutrients 2019, 11, 1613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodall, E.C.A.; Robinson, A.; Johnston, I.G.; Jabbari, S.; Turner, K.A.; Cunningham, A.F.; Lund, P.A.; Cole, J.A.; Henderson, I.R.; Kline, K.A. The Essential Genome of Escherichia coli K-12. MBio 2018, 9, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef] [Green Version]
- Tamanai-Shacoori, Z.; Smida, I.; Bousarghin, L.; Loreal, O.; Meuric, V.; Fong, S.B.; Bonnaure-Mallet, M.; Jolivet-Gougeon, A. Roseburia spp.: A marker of health? Future Microbiol. 2017, 12, 157–170. [Google Scholar] [CrossRef]
- Naaber, P.; Smidt, I.; Štšepetova, J.; Brilene, T.; Annuk, H.; Mikelsaar, M. Inhibition of Clostridium difficile strains by intestinal Lactobacillus species. J. Med. Microbiol. 2004, 53, 551–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreira-Halder, C.V.; de Faria, A.V.S.; Andrade, S.S. Action and function of Faecalibacterium prausnitzii in health and disease. Best Pract. Res. Clin. Gastroenterol. 2017, 31, 643–648. [Google Scholar] [CrossRef] [PubMed]
- Precup, G.; Vodnar, D.-C. Gut Prevotella as a possible biomarker of diet and its eubiotic versus dysbiotic roles: A comprehensive literature review. Br. J. Nutr. 2019, 122, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, A.; Ahmed, A.M.S.; Subramanian, S.; Griffin, N.W.; Drewry, L.L.; Petri, W.A.; Haque, R.; Ahmed, T.; Gordon, J.I. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature 2014, 515, 423–426. [Google Scholar] [CrossRef] [Green Version]
- Vacca, M.; Celano, G.; Calabrese, F.M.; Portincasa, P.; Gobbetti, M.; De Angelis, M. The controversial role of human gut Lachnospiraceae. Microorganisms 2020, 8, 573. [Google Scholar] [CrossRef]
- Grimm, V.; Westermann, C.; Riedel, C.U. Bifidobacteria-Host Interactions—An Update on Colonisation Factors. BioMed Res. Int. 2014, 2014, 960826. [Google Scholar] [CrossRef]
- Hagan, C.L.; Wzorek, J.S.; Kahne, D. Inhibition of the β-barrel assembly machine by a peptide that binds BamD. Proc. Natl. Acad. Sci. USA 2015, 112, 2011–2016. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zhu, X.; Zhang, J.; Lin, Y.; You, X.; Chen, M.; Wang, Y.; Zhu, N.; Si, S. Identification of a Compound That Inhibits the Growth of Gram-Negative Bacteria by Blocking BamA—BamD Interaction. Front. Microbiol. 2020, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Heath, R.; White, S.; Rock, C. Inhibitors of fatty acid synthesis as antimicrobial chemotherapeutics. Appl. Microbiol. Biotechnol. 2002, 58, 695–703. [Google Scholar] [PubMed]
- Pathania, R.; Zlitni, S.; Barker, C.; Das, R.; Gerritsma, D.A.; Lebert, J.; Awuah, E.; Melacini, G.; Capretta, F.A.; Brown, E.D. Chemical genomics in Escherichia coli identifies an inhibitor of bacterial lipoprotein targeting. Nat. Chem. Biol. 2009, 5, 849–856. [Google Scholar] [CrossRef] [PubMed]
- Vetterli, S.U.; Zerbe, K.; Müller, M.; Urfer, M.; Mondal, M.; Wang, S.Y.; Moehle, K.; Zerbe, O.; Vitale, A.; Pessi, G.; et al. Thanatin targets the intermembrane protein complex required for lipopolysaccharide transport in Escherichia coli. Sci. Adv. 2018, 4, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Li, Y.; Wang, W.; Zhang, J.; Lin, Y.; Hong, B.; You, X.; Song, D.; Wang, Y.; Jiang, J.; et al. Identification of an anti-Gram-negative bacteria agent disrupting the interaction between lipopolysaccharide transporters LptA and LptC. Int. J. Antimicrob. Agents 2019, 53, 442–448. [Google Scholar] [CrossRef] [PubMed]
- Urfer, M.; Bogdanovic, J.; Monte, F.L.; Moehle, K.; Zerbe, K.; Omasits, U.; Ahrens, C.H.; Pessi, G.; Eberl, L.; Robinson, J.A. A peptidomimetic antibiotic targets outer membrane proteins and disrupts selectively the outer membrane in Escherichia coli. J. Biol. Chem. 2016, 291, 1921–1932. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Petrushenko, Z.M.; Walker, J.K.; Baudry, J.; Zgurskaya, H.I.; Rybenkov, V.V. Small Molecule Condensin Inhibitors. ACS Infect. Dis. 2018, 4, 1737–1745. [Google Scholar] [CrossRef]
- Tsao, D.H.H.; Sutherland, A.G.; Jennings, L.D.; Li, Y.; Rush, T.S.; Alvarez, J.C.; Ding, W.; Dushin, E.G.; Dushin, R.G.; Haney, S.A.; et al. Discovery of novel inhibitors of the ZipA/FtsZ complex by NMR fragment screening coupled with structure-based design. Bioorganic Med. Chem. 2006, 14, 7953–7961. [Google Scholar] [CrossRef]
- Jennings, L.D.; Foreman, K.W.; Rush, T.S.; Tsao, D.H.H.; Mosyak, L.; Li, Y.; Sukhdeo, M.N.; Ding, W.; Dushin, E.G.; Kenny, C.H.; et al. Design and synthesis of indolo [2,3-a]quinolizin-7-one inhibitors of the ZipA-FtsZ interaction. Bioorg. Med. Chem. Lett. 2004, 14, 1427–1431. [Google Scholar] [CrossRef]
- Ramage, B.; Erolin, R.; Held, K.; Gasper, J.; Weiss, E.; Brittnacher, M.; Gallagher, L.; Manoil, C. Comprehensive Arrayed Transposon Mutant Library of Klebsiella pneumoniae Outbreak Strain KPNIH1. J. Bacteriol. 2017, 199, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Berman, H.M.; Battistuz, T.; Bhat, T.N.; Bluhm, W.F.; Bourne, P.E.; Burkhardt, K.; Feng, Z.; Gilliland, G.L.; Iype, L.; Jain, S.; et al. The protein data bank. Acta Crystallogr. Sect. D Biol. Crystallogr. 2002, 58, 899–907. [Google Scholar] [CrossRef]
- Ramos, P.I.P.; Fernández Do Porto, D.; Lanzarotti, E.; Sosa, E.J.; Burguener, G.; Pardo, A.M.; Klein, C.C.; Sagot, M.; de Vasconcelos, A.T.R.; Gales, A.C.; et al. An integrative, multi-omics approach towards the prioritization of Klebsiella pneumoniae drug targets. Sci. Rep. 2018, 8, 10755. [Google Scholar] [CrossRef] [Green Version]
- Hadizadeh, M.; Tabatabaiepour, S.N.; Tabatabaiepour, S.Z.; Hosseini Nave, H.; Mohammadi, M.; Sohrabi, S.M. Genome-Wide Identification of Potential Drug Target in Enterobacteriaceae Family: A Homology-Based Method. Microb. Drug Resist. 2018, 24, 8–17. [Google Scholar] [CrossRef]
- Georrge, J.J.; Umrania, V. In silico identification of putative drug targets in Klebsiella pneumonia MGH78578. Indian J. Biotechnol. 2011, 10, 432–439. [Google Scholar]
- Duffield, M.; Cooper, I.; McAlister, E.; Bayliss, M.; Ford, D.; Oyston, P. Predicting conserved essential genes in bacteria: In silico identification of putative drug targets. Mol. Biosyst. 2010, 6, 2482. [Google Scholar] [CrossRef] [PubMed]
- Mondal, S.I.; Ferdous, S.; Akter, A.; Mahmud, Z.; Karim, N.; Islam, M.M.; Jewel, N.A.; Afrin, T. Identification of potential drug targets by subtractive genome analysis of Escherichia coli O157:H7: An in silico approach. Adv. Appl. Bioinforma. Chem. 2015, 8, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grabowicz, M. Lipoprotein Transport: Greasing the Machines of Outer Membrane Biogenesis. BioEssays 2018, 40, 1700187. [Google Scholar] [CrossRef] [PubMed]
- Lycklama A Nijeholt, J.A.; De Keyzer, J.; Prabudiansyah, I.; Driessen, A.J.M. Characterization of the supporting role of SecE in protein translocation. FEBS Lett. 2013, 587, 3083–3088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein, G.; Kobylak, N.; Lindner, B.; Stupak, A.; Raina, S. Assembly of Lipopolysaccharide in Escherichia coli Requires the Essential LapB Heat Shock Protein. J. Biol. Chem. 2014, 289, 14829–14853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.Q.; Dixon, N.E. Bacterial replisomes. Curr. Opin. Struct. Biol. 2018, 53, 159–168. [Google Scholar] [CrossRef]
- Huang, Y.H.; Huang, C.Y. Structural insight into the DNA-binding mode of the primosomal proteins PriA, PriB, and AnaT. BioMed Res. Int. 2014, 2014, 195162. [Google Scholar] [CrossRef] [Green Version]
- Ote, T.; Hashimoto, M.; Ikeuchi, Y.; Su’etsugu, M.; Suzuki, T.; Katayama, T.; Kato, J.I. Involvement of the Escherichia coli folate-binding protein YgfZ in RNA modification and regulation of chromosomal replication initiation. Mol. Microbiol. 2006, 59, 265–275. [Google Scholar] [CrossRef]
- Rybenkov, V.V.; Herrera, V.; Petrushenko, Z.M.; Zhao, H. MukBEF, a Chromosomal Organizer. J. Mol. Microbiol. Biotechnol. 2014, 24, 371–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Condon, S.G.F.; Mahbuba, D.A.; Armstrong, C.R.; Diaz-Vazquez, G.; Craven, S.J.; LaPointe, L.M.; Khadria, A.S.; Chadda, R.; Crooks, J.A.; Rangarajan, N.; et al. The FtsLB subcomplex of the bacterial divisome is a tetramer with an uninterrupted FtsL helix linking the transmembrane and periplasmic regions. J. Biol. Chem. 2018, 293, 1623–1641. [Google Scholar] [CrossRef] [Green Version]
- Buddelmeijer, N.; Beckwith, J. A complex of the Escherichia coli cell division proteins FtsL, FtsB and FtsQ forms independently of its localization to the septal region. Mol. Microbiol. 2004, 52, 1315–1327. [Google Scholar] [CrossRef] [PubMed]
- Hale, C.A.; De Boer, P.A.J. ZipA is required for recruitment of FtsK, FtsQ, FtsL, and FtsN to the septal ring in Escherichia coli. J. Bacteriol. 2002, 184, 2552–2556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kruse, T.; Bork-Jensen, J.; Gerdes, K. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol. Microbiol. 2005, 55, 78–89. [Google Scholar] [CrossRef]
- Ikeuchi, Y.; Shigi, N.; Kato, J.I.; Nishimura, A.; Suzuki, T. Mechanistic insights into sulfur relay by multiple sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Mol. Cell 2006, 21, 97–108. [Google Scholar] [CrossRef]
- Vanorsdel, C.E.; Bhatt, S.; Allen, R.J.; Brenner, E.P.; Hobson, J.J.; Jamil, A.; Haynes, B.M.; Genson, A.M.; Hemma, M.R. The Escherichia coli CydX protein is a member of the CydAB cytochrome bd oxidase complex and is required for cytochrome bd oxidase activity. J. Bacteriol. 2013, 195, 3640–3650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bougdour, A.; Cunning, C.; Baptiste, P.J.; Elliott, T.; Gottesman, S. Multiple pathways for regulation of σS (RpoS) stability in Escherichia coli via the action of multiple anti-adaptors. Mol. Microbiol. 2008, 68, 298–313. [Google Scholar] [CrossRef] [PubMed]
- Ishii, E.; Eguchi, Y.; Utsumi, R. Mechanism of activation of PhoQ/PhoP two-component signal transduction by SafA, an auxiliary protein of PhoQ histidine kinase in Escherichia coli. Biosci. Biotechnol. Biochem. 2013, 77, 814–819. [Google Scholar] [CrossRef]
- Kajimura, J.; Rahman, A.; Rick, P.D. Assembly of cyclic enterobacterial common antigen in Escherichia coli K-12. J. Bacteriol. 2005, 187, 6917–6927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heath, R.J.; Rock, C.O. Roles of the FabA and FabZ β-hydroxyacyl-acyl carrier protein dehydratases in Escherichia coli fatty acid biosynthesis. J. Biol. Chem. 1996, 271, 27795–27801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zurawski, G.; Elseviers, D.; Stauffer, G.V.; Yanofsky, C. Translational control of transcription termination at the attenuator of the Escherichia coli tryptophan operon. Proc. Natl. Acad. Sci. USA 1978, 75, 5988–5992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jordan, P.M.; Mgbeje, B.I.; Thomas, S.D.; Alwan, A.F. Nucleotide sequence for the hemD gene of Escherichia coli encoding uroporphyrinogen III synthase and initial evidence for a hem operon. Biochem. J. 1988, 249, 613–616. [Google Scholar] [CrossRef] [Green Version]
- Fraikin, N.; Goormaghtigh, F.; Van Melderen, L. Type II Toxin-Antitoxin Systems: Evolution and Revolutions. J. Bacteriol. 2020, 202, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Springer, M.; Mayaux, J.F.; Fayat, G.; Plumbridge, J.A.; Graffe, M.; Blanquet, S.; Grunberg-Manago, M. Attenuation control of the Escherichia coli phenylalanyl-tRNA synthetase operon. J. Mol. Biol. 1985, 181, 467–478. [Google Scholar] [CrossRef]
- Merino, E.; Jensen, R.A.; Yanofsky, C. Evolution of bacterial trp operons and their regulation. Curr. Opin. Microbiol. 2008, 11, 78–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melior, H.; Maaß, S.; Li, S.; Förstner, K.U.; Azarderakhsh, S.; Varadarajan, A.R.; Stötzel, M.; Elhossary, M.; Barth-Weber, S.; Ahrens, C.H.; et al. The leader peptide peTrpL forms antibiotic-containing ribonucleoprotein complexes for posttranscriptional regulation of multiresistance genes. MBio 2020, 11, 1–22. [Google Scholar] [CrossRef]
- Langdon, A.; Crook, N.; Dantas, G. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med. 2016, 8, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wyres, K.L.; Lam, M.M.C.; Holt, K.E. Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 2020, 18, 344–359. [Google Scholar] [CrossRef] [PubMed]
- Silver, L.L. Appropriate Targets for Antibacterial Drugs. Cold Spring Harb. Perspect. Med. 2016, 6, a030239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mobegi, F.M.; van Hijum, S.A.F.T.; Burghout, P.; Bootsma, H.J.; de Vries, S.P.W.; Jongh, C.E.; Simonetti, E.; Langereis, J.D.; Hermans, P.W.M.; de Jonge, M.I.; et al. From microbial gene essentiality to novel antimicrobial drug targets. BMC Genom. 2014, 15, 958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakheet, T.M.; Doig, A.J. Properties and identification of antibiotic drug targets. BMC Bioinform. 2010, 11, 195. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Gene | Protein Function | PDB Accession No. | Crystal Structure and Resolution | SCL | Known Inhibitor | Conservation and Essentiality in K. pneumoniae | |||
|---|---|---|---|---|---|---|---|---|---|
| Alignment | aa Identity | Bitscore | Essentiality Status | ||||||
| bamD | Outer membrane protein assembly factor BamD | P0AC02 | 5D0O 2.90 Å | Cell outer membrane; Lipid anchor | Inhibitory peptide [19], IMB-H4 [20] | 100% | 94% | 479 | Essential |
| cydX | Cytochrome bd-I ubiquinol oxidase subunit X | P56100 | 6RKO 2.68 Å | Cell inner membrane; Single-pass membrane protein | N/A | 100% | 100% | 77 | N/A |
| dnaT | Primosomal protein 1 | P0A8J2 | 4OU6 1.96 Å | N/D | N/A | 100% | 76% | 285 | N/A |
| fabA | 3-hydroxydecanoyl-[acyl-carrier-protein] dehydratase | P0A6Q3 | 4KEH 2Å | Cytoplasm | 3-Decynoyl-NAC [21] | 100% | 99% | 349 | Essential |
| ftsB | Cell division protein FtsB | P0A6S5 | 4IFF 2.30 Å | Cell inner membrane | N/A | 84% | 100% | 181 | Essential |
| ftsL | Cell division protein FtsL | P0AEN4 | N/D | Cell inner membrane | N/A | 100% | 96% | 238 | Essential |
| ftsQ | Cell division protein FtsQ | P06136 | 2VH1 2.70 Å | Cell inner membrane; Single-pass type II membrane protein | N/A | 96% | 89% | 486 | Essential |
| hemD | Uroporphyrinogen-III synthase | P09126 | N/D | N/D | N/A | 100% | 99% | 497 | Essential |
| higA | Antitoxin HigA | P67701 | 6JQ4 2 Å | N/D | N/A | 100% | 99% | 279 | N/A |
| hipB | Antitoxin HipB | P23873 | 2WIU 2.35 Å | N/D | N/A | 100% | 98% | 176 | Essential |
| holD | DNA polymerase III subunit psi | P28632 | 3SXU 1.85 Å | N/D | N/A | 84% | 98% | 224 | N/A |
| iraM | Anti-adapter protein IraM | P75987 | N/D | Cytoplasm | N/A | 99% | 62% | 149 | N/A |
| lolA | Outer membrane lipoprotein carrier protein | P61316 | 1IWL 1.56 Å | Periplasm | MAC13243 [22] | 100% | 94% | 399 | Essential |
| lolB | Outer membrane lipoprotein LolB | P61320 | 1IWM 1.90 Å | Cell outer membrane; Lipid anchor | N/A | 100% | 100% | 426 | Essential |
| lptA | Lipopolysaccharide export system protein LptA | P0ADV1 | 2R19 2.16 Å | Periplasm | Thanatin [23], Compound IMB-881 [24] | 100% | 85% | 328 | Essential |
| lptD | LPS assembly protein LptD | P31554 | 4RHB 3.35 Å | Cell outer membrane | Inhibitory peptide JB-95 [25] | 100% | 84% | 1407 | Essential |
| lptE | LPS assembly lipoprotein LptE | P0ADC1 | 4RHB 3.35 Å | Cell outer membrane | N/A | 102% | 72% | 295 | Essential |
| mreD | Rod shape-determining protein MreD | P0ABH4 | N/D | Cell inner membrane; Multi-pass membrane protein | N/A | 100% | 100% | 310 | N/A |
| mukB | Chromosome partition protein MukB | P22523 | 1QHL 2.20 Å | Nucleoid | NSC260594, Michellamine B [26] | 100% | 92% | 2774 | Essential |
| mukE | Chromosome partition protein MukE | P22524 | 3EUH 2.90 Å | Nucleoid | N/A | 100% | 95% | 458 | Essential |
| mukF | Chromosome partition protein MukF | P60293 | 3EUH 2.90 Å | Nucleoid | N/A | 100% | 97% | 878 | Essential |
| pheM | Phenylalanine tRNA ligase operon leader peptide | P0AD74 | N/D | N/D | N/A | 100% | 100% | 30 | N/A |
| priB | Primosomal replication protein N | P07013 | 5WQV 1.97 Å | N/D | N/A | 100% | 93% | 205 | N/A |
| safA | Two-component-system connector protein SafA | P76136 | N/D | Cell inner membrane; Single-pass type II membrane protein | N/A | 98% | 100% | 127 | N/A |
| secE | Protein translocase subunit SecE | P0AG96 | 5GAE 3.33 Å | Cell inner membrane; Multi-pass membrane protein | N/A | 100% | 98% | 240 | Essential |
| trpL | trp operon leader peptide | P0AD92 | N/D | N/D | N/A | 100% | 100% | 32 | N/A |
| tusE | Sulfurtransferase TusE | P0AB18 | N/D | Cytoplasm | N/A | 100% | 90% | 204 | Essential |
| wzyE | Probable ECA polymerase | P27835 | N/D | Cell inner membrane | N/A | 100% | 87% | 744 | N/A |
| ycaR | UPF0434 protein YcaR | P0AAZ7 | N/D | Cytoplasm | N/A | 100% | 100% | 125 | N/A |
| yciS | Lipopolysaccharide assembly protein A | P0ACV4 | N/D | Cell inner membrane | N/A | 100% | 87% | 177 | N/A |
| ydfO | Uncharacterized protein YdfO | P76156 | 2HH8 N/D | N/D | N/A | 100% | 79% | 228 | N/A |
| ydhL | Uncharacterized protein YdhL | P64474 | N/D | N/D | N/A | 96% | 98% | 152 | N/A |
| ygfZ | tRNA-modifying protein YgfZ | P0ADE8 | 1VLY 1.30 Å | Cytoplasm | N/A | 100% | 84% | 573 | N/A |
| yqeL | Uncharacterized protein YqeL | C1P613 | N/D | N/D | N/A | N/A | N/A | N/A | N/A |
| yrfF | Putative membrane protein IgaA homolog | P45800 | 4UZM N/D | Cell inner membrane; Multi-pass membrane protein | N/A | 100% | 73% | 1089 | N/A |
| zipA | Cell division protein ZipA | P77173 | 1F46 1.50 Å | Cell inner membrane; Single-pass type I membrane protein | Antimicrobial compounds [27,28] | 101% | 98% | 646 | Essential |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Svanberg Frisinger, F.; Jana, B.; Donadio, S.; Guardabassi, L. In Silico Prediction and Prioritization of Novel Selective Antimicrobial Drug Targets in Escherichia coli. Antibiotics 2021, 10, 632. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10060632
Svanberg Frisinger F, Jana B, Donadio S, Guardabassi L. In Silico Prediction and Prioritization of Novel Selective Antimicrobial Drug Targets in Escherichia coli. Antibiotics. 2021; 10(6):632. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10060632
Chicago/Turabian StyleSvanberg Frisinger, Frida, Bimal Jana, Stefano Donadio, and Luca Guardabassi. 2021. "In Silico Prediction and Prioritization of Novel Selective Antimicrobial Drug Targets in Escherichia coli" Antibiotics 10, no. 6: 632. https://0-doi-org.brum.beds.ac.uk/10.3390/antibiotics10060632






