Development of a Natural Preservative from Chestnut Flowers: Ultrasound-Assisted Extraction Optimization and Functionality Assessment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Optimization of the Extraction Process to Obtain an Extract Rich in Phenolic Compounds, from Male Flowers of Castanea sativa Mill
2.3. Ultrasound-Assisted Extractions
2.4. Preparation of Extracts Obtained by Ultrasound-Assisted Extraction
2.5. Identification and Quantification of Phenolic Compounds by HPLC-DAD-ESI/MS
2.6. Experimental Design, Modeling, and Optimization
2.6.1. Mathematical Model
2.6.2. Procedure for Optimization of Variables
2.6.3. Numerical Methods, Statistical Analysis, and Graphic Illustrations
2.7. Bioactivities of the Optimized Extract from Chestnut Male Flower
2.7.1. Extract’s Preparation
2.7.2. Evaluation of Antioxidant Activity
2.7.3. Evaluation of Anti-Inflammatory Activity
2.7.4. Evaluation of the Cytotoxic and Hepatotoxic Activity
2.7.5. Evaluation of Antimicrobial Activity
3. Results and Discussion
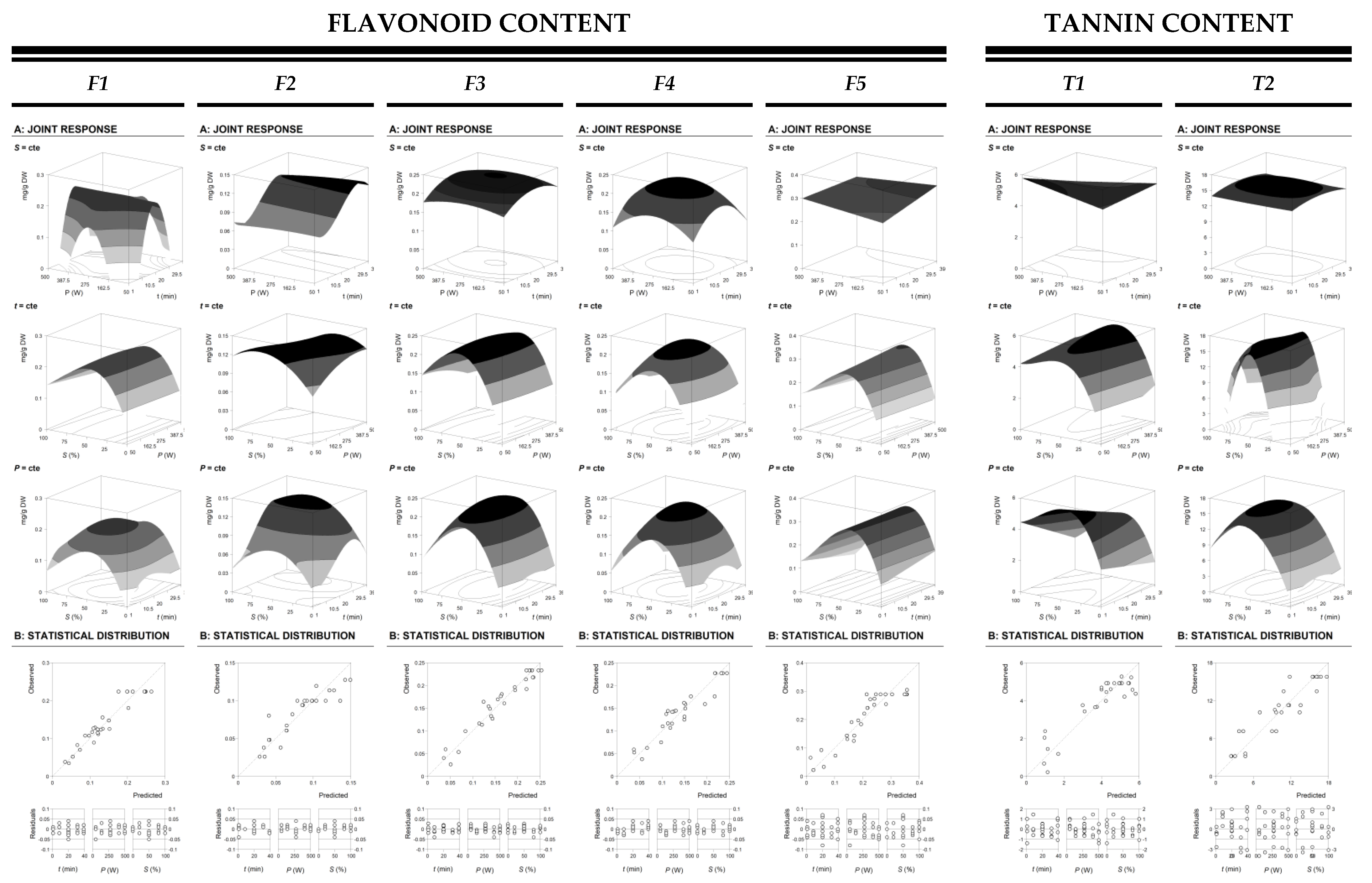
3.1. Theoretical Response Surface Models of the Used Response Criteria for the RSM Analysis and Statistical Verification
3.2. Effect of the Extraction Variables on the Target Responses
3.3. Numerical Optimal Conditions That Maximize the Extraction and Experimental Verification of Predictive Models
- Regarding flavonoids, individual variable conditions were found for F1 at 17.64 ± 2.59 min, 291.45 ± 17.07 W and 43.28 ± 6.58% of ethanol, producing maximum response values of 0.23 ± 0.13 mg F1/g R. For F2, variable conditions at 33.06 ± 3.46 min, 50.00 ± 7.07 W and 65.80 ± 8.11% of ethanol were found to produce a maximum response values of 0.13 ± 0.04 mg F2/g R. In the case of F3, variable conditions were found at 23.37 ± 2.94 min, 275.00 ± 16.58 W and 50.00 ± 7.07% of ethanol, producing a maximum response values of 0.23 ± 0.05 mg F3/g R. Variable conditions at 20.11 ± 2.75 min, 279.01 ± 16.70 W and 45.20 ± 6.72% of ethanol were found to produce a maximum response values of 0.23 ± 0.05 mg F4/g R in the case of F4. Variable conditions were found for F5 at 9.31 ± 1.96 min, 500.00 ± 22.36 W and 46.12 ± 6.79% of ethanol for F5, producing maximum response values of 0.30 ± 0.05 mg F5/g R. At last, for total flavonoids, individual variable conditions at 23.92 ± 2.97 min, 289.36 ± 17.01 W and 44.18 ± 6.65% of ethanol were found to produce maximum response values of 1.12 ± 0.08 mg FT/g R.
- In respect to tannins, individual variable conditions were found at 1.00 ± 0.24 min. 500.00 ± 22.36 W and 46.31 ± 6.81% of ethanol, producing maximum response values of 5.78 ± 0.39 mg T1/g R for T1 whereas individual variable conditions at 20.00 ± 2.74 min, 275.00 ± 16.58 W and 59.96 ± 7.74% of ethanol were found to produce maximum response values of 16.13 ± 0.90 mg in the case of T2/g R. Finally, for total tannins content, 8.52 ± 1.89 min, 500.00 ± 22.36 W and 52.71 ± 7.26% of ethanol were the individual variable conditions found to be able to produce a maximum response values of 23.35 ± 0.47 mg TT/g R.
- Regarding the relative optimal response values for flavonoid content, the global optimal variable conditions were found at 26.32 ± 3.07 min, 285.57 ± 16.90 W and 44.80 ± 6.69% of ethanol, achieving maximum response values of 0.21 ± 0.06 mg F1/g E, 0.12 ± 0.04 mg F2/g R, 0.23 ± 0.08 mg F3/g R, 0.22 ± 0.07 mg F4/g R, 0.30 ± 0.05 mg F5/g R, respectively, from F1 to F5 compounds. In the case of tannins under optimal response values for flavonoid content, their maximum response values were 4.79 ± 0.19 mg T1 /g R and 15.12 ± 1.89 mg T2/g R, respectively, for each tannin. Focusing on the yield of the extracted residue, it was lower than in the case of optimizing individual variables, but still reaches 0.13 ± 0.07 g R/g DW.
- In respect to the relative optimal response values for tannin content, global optimal variable conditions at 11.44 ± 2.03 min, 500.00 ± 22.39 W and 51.22 ± 7.16% of ethanol were found to produce maximum response levels of 5.35 ± 0.31 mg T1/g R and 15.23 ± 0.90 mg T2/g R, respectively. In the case of flavonoids, responses values ranged from 0.07 ± 0.01 mg F1/g R, 0.07 ± 0.02 mg F2/g R, 0.20 ± 0.05 mg F3/g R, 0.16 ± 0.03 mg F4/g R, and 0.29 ± 0.04 mg F5/g R. Furthermore, the maximum yield obtained at these fixed conditions was 0.15 g R/g DW.
- At the end, the global optimal variable conditions for the optimization of both, flavonoids and tannins content and also yield, were found at 23.47 ± 2.90 min, 258.78 ± 16.09 W and 50.51 ± 7.11% of ethanol, thus producing the following maximum response values: 0.13 ± 0.07 g R/ g DW, 0.22 ± 0.07 mg F1/g R, 0.11 ± 0.03 mg F2/g R, 0.23 ± 0.08 mg F3/g R, 0.23 ± 0.07 mg F4/g R, 0.29 ± 0.04 mg F5/g R, 4.88 ± 0.21 mg T1/g R, and 15.77 ± 0.97 mg T2/g R.
3.4. Evaluation of Bioactivities of Chestnut Flowers Optimized Extract
3.4.1. Antioxidant Activity
3.4.2. Anti-Inflammatory and Cytotoxic Properties in Cell Lines
3.4.3. Antimicrobial Activity
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Socaci, S.A.; Fărcaş, A.C.; Tofană, M. Functional ingredients derived from aromatic plants. Feed Addit. 2020, 133–146. [Google Scholar] [CrossRef]
- Munekata, P.E.; Pateiro, M.; Bellucci, E.R.B.; Domínguez, R.; Barretto, A.C.D.S.; Lorenzo, J.M. Strategies to increase the shelf life of meat and meat products with phenolic compounds. Mar. Med. Foods Implic. Appl. Anim. Microbes 2021. [Google Scholar] [CrossRef]
- Foo, J.; Michor, F. Evolution of resistance to anti-cancer therapy during general dosing schedules. J. Theor. Biol. 2010, 263, 179–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carocho, M.; Barros, L.; Bento, A.; Santos-Buelga, C.; Morales, P.; Ferreira, I.C.F.R. Castanea sativa mill. Flowers amongst the Most Powerful Antioxidant Matrices: A Phytochemical Approach in Decoctions and Infusions. BioMed Res. Int. 2014, 2014, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caleja, C.; Barros, L.; Prieto, M.A.; Bento, A.; Oliveira, M.B.P.P.; Ferreira, I.C.F.R. Development of a natural preservative obtained from male chestnut flowers: Optimization of a heat-assisted extraction technique. Food Funct. 2019, 10, 1352–1363. [Google Scholar] [CrossRef]
- Carocho, M.; Barreira, J.C.; Barros, L.; Bento, A.; Cámara, M.; Morales, P.; Ferreira, I.C. Traditional pastry with chestnut flowers as natural ingredients: An approach of the effects on nutritional value and chemical composition. J. Food Compos. Anal. 2015, 44, 93–101. [Google Scholar] [CrossRef] [Green Version]
- Chemat, F.; Rombaut, N.; Sicaire, A.G.; Meullemiestre, A.; Fabiano-Tixier, A.S.; Abert-Vian, M. Ultrasound assisted extrac-tion of food and natural products. Mechanisms, techniques, combinations, protocols and applications. A review. Ultrason. Sonochem. 2017, 34, 540–560. [Google Scholar] [CrossRef] [PubMed]
- Diouf, P.N.; Stevanovic, T.; Boutin, Y. The effect of extraction process on polyphenol content, triterpene composition and bioactivity of yellow birch (Betula alleghaniensis Britton) extracts. Ind. Crop. Prod. 2009, 30, 297–303. [Google Scholar] [CrossRef]
- Chavan, Y.; Singhal, R.S. Ultrasound-assisted extraction (UAE) of bioactives from arecanut (Areca catechu L.) and optimization study using response surface methodology. Innov. Food Sci. Emerg. Technol. 2013, 17, 106–113. [Google Scholar] [CrossRef]
- Mandpe, S.R.; Parate, V.R.; Naik, J.B. Method optimization and analysis of flurbiprofen loaded Eudragit L100 nanoparticles using RP-HPLC technique: A central composite design approach. Mater. Today Proc. 2021, 1–10. [Google Scholar] [CrossRef]
- López, C.J.; Caleja, C.; Prieto, M.A.; Barreiro, M.F.; Barros, L.; Ferreira, I.C. Optimization and comparison of heat and ultrasound assisted extraction techniques to obtain anthocyanin compounds from Arbutus unedo L. Fruits. Food Chem. 2018, 264, 81–91. [Google Scholar] [CrossRef] [Green Version]
- Bessada, S.M.; Barreira, J.C.; Barros, L.; Ferreira, I.C.; Oliveira, M.B.P. Phenolic profile and antioxidant activity of Coleostephus myconis (L.) Rchb.f.: An underexploited and highly disseminated species. Ind. Crop. Prod. 2016, 89, 45–51. [Google Scholar] [CrossRef] [Green Version]
- Vieira, V.; Prieto, M.A.; Barros, L.; Coutinho, J.A.P.; Ferreira, O.; Ferreira, I.C.F.R. Optimization and comparison of macera-tion and microwave extraction systems for the production of phenolic compounds from Juglans regia L. for the valorization of walnut leaves. Ind. Crop. Prod. 2017, 107, 341–352. [Google Scholar] [CrossRef] [Green Version]
- Prieto, M.; Vázquez, J.; Murado, M. A new mathematical model to quantify and characterize the response to pro- and anti-oxidants of the copper-induced oxidation of LDL assay. A tool for examination of potential preventive compounds and clinical risk prediction. Food Res. Int. 2014, 66, 501–513. [Google Scholar] [CrossRef] [Green Version]
- Murado, M.A.; Prieto, M.A. Dose-Response Analysis in the Joint Action of Two Effectors. A New Approach to Simulation, Identification and Modelling of Some Basic Interactions. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [Green Version]
- Hodges, D.M.; Delong, J.M.; Forney, C.F.; Prange, R.K. Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 1999, 207, 604–611. [Google Scholar] [CrossRef]
- Lockowandt, L.; Pinela, J.; Roriz, C.L.; Pereira, C.; Abreu, R.M.; Calhelha, R.C.; Alves, M.J.; Barros, L.; Bredol, M.; Ferreira, I.C. Chemical features and bioactivities of cornflower (Centaurea cyanus L.) capitula: The blue flowers and the unexplored non-edible part. Ind. Crop. Prod. 2019, 128, 496–503. [Google Scholar] [CrossRef] [Green Version]
- Svobodova, B.; Barros, L.; Sopik, T.; Calhelha, R.C.; Heleno, S.; Alves, M.J.; Walcott, S.; Kuban, V.; Ferreira, I.C.F.R. Non-edible parts of Solanum stramoniifolium Jacq.—A new potent source of bioactive extracts rich in phenolic compounds for functional foods. Food Funct. 2017, 8, 2013–2021. [Google Scholar] [CrossRef] [Green Version]
- Guimarães, R.; Calhelha, R.C.; Froufe, H.J.C.; Abreu, R.M.V.; Carvalho, A.M.; João, R.P.Q.M.; Ferreira, I.C.F.R. Wild Roman chamomile extracts and phenolic compounds: Enzymatic assays and molecular modelling studies with VEGFR-2 tyrosine kinase. Food Funct. 2015, 7, 79–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soković, M.; Glamoćlija, J.; Marin, M.D.; Brkić, D.; van Griensven, L.J.L.D. Antibacterial effects of the essential oils of com-monly consumed medicinal herbs using an in vitro model. Molecules 2010, 15, 7532–7546. [Google Scholar] [CrossRef] [Green Version]
- Soković, M.; van Griensven, L.J.L.D. Antimicrobial activity of essential oils and their components against the three major pathogens of the cultivated button mushroom, Agaricus bisporus. Eur. J. Plant Pathol. 2006, 116, 211–224. [Google Scholar] [CrossRef]
- Pinela, J.; Prieto, M.A.; Carvalho, A.M.; Barreiro, M.F.; Oliveira, M.B.P.; Barros, L.; Ferreira, I.C. Microwave-assisted extraction of phenolic acids and flavonoids and production of antioxidant ingredients from tomato: A nutraceutical-oriented optimization study. Sep. Purif. Technol. 2016, 164, 114–124. [Google Scholar] [CrossRef] [Green Version]
- Caleja, C.; Barros, L.; Barreira, J.C.M.; Soković, M.; Calhelha, R.C.; Bento, A.; Oliveira, M.B.P.P.; Ferreira, I.C.F.R. Castanea sativa male flower extracts as an alternative additive in the Portuguese pastry delicacy “pastel de nata”. Food Funct. 2020, 11, 2208–2217. [Google Scholar] [CrossRef] [PubMed]
- Bezerra, M.A.; Santelli, R.E.; Oliveira, E.P.; Villar, L.S.; Escaleira, E.A.; Escaleira, L.A. Response surface methodology (RSM) as a tool for optimization in analytical chemistry. Talanta 2008, 76, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Dias, M.I.; Ferreira, I.C.F.R.; Barreiro, M.F. Microencapsulation of bioactives for food applications. Food Funct. 2015, 6, 1035–1052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Echegaray, N.; Gómez, B.; Barba, F.J.; Franco, D.; Estévez, M.; Carballo, J.; Marszałek, K.; Lorenzo, J.M. Chestnuts and by-products as source of natural antioxidants in meat and meat products: A review. Trends Food Sci. Technol. 2018, 82, 110–121. [Google Scholar] [CrossRef]
- Antonio, A.L.; Pereira, E.; Pinela, J.; Heleno, S.A.; Pereira, C.; Ferreira, I.C. Determination of Antioxidant Compounds in Foodstuff. In Food Safety: Innovative Analytical Tools for Safety Assessment; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2016; pp. 179–220. [Google Scholar]
- Misra, N.N.; Martynenko, A.; Chemat, F.; Paniwnyk, L.; Barba, F.J.; Jambrak, A.R. Thermodynamics, transport phenomena, and electrochemistry of external field-assisted nonthermal food technologies. Crit. Rev. Food Sci. Nutr. 2018, 58, 1832–1863. [Google Scholar] [CrossRef] [Green Version]
- Tomšik, A.; Pavlić, B.; Vladić, J.; Ramić, M.; Brindza, J.; Vidović, S. Optimization of ultrasound-assisted extraction of bioac-tive compounds from wild garlic (Allium ursinum L.). J. Food Sci. Technol. 2020, 57, 4627–4636. [Google Scholar]
- Montesano, D.; Fallarino, F.; Cossignani, L.; Bosi, A.; Simonetti, M.S.; Puccetti, P.; Damiani, P. Innovative extraction proce-dure for obtaining high pure lycopene from tomato. Eur. Food Res. Technol. 2008, 226, 327–335. [Google Scholar] [CrossRef]
- Zhu, Z.; He, J.; Liu, G.; Barba, F.J.; Koubaa, M.; Ding, L.; Bals, O.; Grimi, N.; Vorobiev, E. Recent insights for the green reco-very of inulin from plant food materials using non-conventional extraction technologies: A review. Innov. Food Sci. Emerg. Technol. 2016, 33, 1–9. [Google Scholar] [CrossRef]
- Engler, M.B.; Engler, M.M. The Emerging Role of Flavonoid-Rich Cocoa and Chocolate in Cardiovascular Health and Disease. Nutr. Rev. 2006, 64, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Nichenametla, S.N.; Taruscio, T.G.; Barney, D.L.; Exon, J.H. A Review of the Effects and Mechanisms of Polyphenolics in Cancer. Crit. Rev. Food Sci. Nutr. 2006, 46, 161–183. [Google Scholar] [CrossRef] [PubMed]
- Lau, F.C.; Shukitt-Hale, B.; Joseph, J.A. The beneficial effects of fruit polyphenols on brain aging. Neurobiol. Aging 2005, 26, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Weaver, C.M.; Cheong, J.M.K. Soy Isoflavones and Bone Health: The Relationship Is Still Unclear. J. Nutr. 2005, 135, 1243–1247. [Google Scholar] [CrossRef] [Green Version]
- Barreira, J.C.; Ferreira, I.C.; Oliveira, M.B.P.; Pereira, J.A. Antioxidant activity and bioactive compounds of ten Portuguese regional and commercial almond cultivars. Food Chem. Toxicol. 2008, 46, 2230–2235. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, L.; Kim, S.H.; Hagerman, A.E.; Lü, J. Anti-cancer, anti-diabetic and other pharmacologic and biological activities of penta-galloyl-glucose. Pharm. Res. 2009, 26, 2066–2080. [Google Scholar] [CrossRef] [Green Version]
- Sanches-Silva, A.; Testai, L.; Nabavi, S.F.; Battino, M.; Devi, K.P.; Tejada, S.; Sureda, A.; Xu, S.; Yousefi, B.; Majidinia, M.; et al. Therapeutic potential of polyphenols in cardiovascular diseases: Regulation of mTOR signaling pathway. Pharmacol. Res. 2020, 152. [Google Scholar] [CrossRef] [PubMed]


| CODED VALUES | NATURAL VALUES | ||
|---|---|---|---|
| t (min) | P (W) | S (%) | |
| −1.68 | 1.0 | 50.0 | 0.0 |
| −1 | 8.7 | 141.2 | 20.3 |
| 0 | 20.0 | 275.0 | 50.0 |
| +1 | 31.3 | 408.8 | 79.7 |
| +1.68 | 39.0 | 500.0 | 100.0 |
| EXPERIMENTAL DESIGN VALUES | RESIDUE | FLAVONOID CONTENT | TANNIN CONTENT | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CODED | NATURAL | INDIVIDUAL | TOTAL | INDIVIDUAL | TOTAL | |||||||||||
| X1 | X2 | X3 | X1: t | X2: P | X3: S | Yield | F1 | F2 | F3 | F4 | F5 | Ft | T1 | T2 | Tt | |
| min | W | % | g R/g DW | mg/g R | mg/g R | mg/g R | mg/g R | mg/g R | mg/g R | mg/g R | mg/g R | mg/g R | ||||
| 1 | −1 | −1 | −1 | 8.7 | 141.2 | 20.3 | 0.10 | 0.15 | 0.06 | 0.14 | 0.13 | 0.21 | 0.69 | 3.67 | 9.67 | 13.34 |
| 2 | 1 | −1 | −1 | 31.3 | 141.2 | 20.3 | 0.10 | 0.09 | 0.12 | 0.14 | 0.12 | 0.22 | 0.68 | 3.10 | 3.73 | 6.83 |
| 3 | −1 | 1 | −1 | 8.7 | 408.8 | 20.3 | 0.12 | 0.11 | 0.06 | 0.17 | 0.15 | 0.28 | 0.78 | 5.82 | 9.00 | 14.82 |
| 4 | 1 | 1 | −1 | 31.3 | 408.8 | 20.3 | 0.12 | 0.13 | 0.14 | 0.16 | 0.15 | 0.23 | 0.82 | 3.00 | 4.41 | 7.41 |
| 5 | −1 | −1 | 1 | 8.7 | 141.2 | 79.7 | 0.11 | 0.11 | 0.06 | 0.16 | 0.11 | 0.18 | 0.63 | 4.49 | 11.75 | 16.24 |
| 6 | 1 | −1 | 1 | 31.3 | 141.2 | 79.7 | 0.10 | 0.10 | 0.15 | 0.19 | 0.13 | 0.22 | 0.79 | 4.00 | 10.06 | 14.06 |
| 7 | −1 | 1 | 1 | 8.7 | 408.8 | 79.7 | 0.17 | 0.13 | 0.06 | 0.12 | 0.11 | 0.16 | 0.59 | 4.00 | 13.57 | 17.57 |
| 8 | 1 | 1 | 1 | 31.3 | 408.8 | 79.7 | 0.16 | 0.11 | 0.13 | 0.16 | 0.15 | 0.19 | 0.74 | 3.75 | 11.57 | 15.32 |
| 9 | 0 | 0 | −1.68 | 20 | 275 | 0 | 0.11 | 0.12 | 0.07 | 0.14 | 0.12 | 0.17 | 0.63 | 4.24 | 9.43 | 13.67 |
| 10 | 0 | 0 | 1.68 | 20 | 275 | 100 | 0.10 | 0.07 | 0.07 | 0.07 | 0.10 | 0.10 | 0.42 | 1.70 | 4.74 | 6.44 |
| 11 | 0 | −1.68 | 0 | 20 | 50 | 50 | 0.11 | 0.18 | 0.09 | 0.23 | 0.16 | 0.21 | 0.88 | 4.94 | 11.82 | 16.76 |
| 12 | 0 | 1.68 | 0 | 20 | 500 | 50 | 0.16 | 0.25 | 0.12 | 0.24 | 0.22 | 0.26 | 1.08 | 5.42 | 18.73 | 24.15 |
| 13 | −1.68 | 0 | 0 | 1 | 275 | 50 | 0.10 | 0.13 | 0.04 | 0.22 | 0.15 | 0.24 | 0.79 | 5.07 | 10.85 | 15.92 |
| 14 | 1.68 | 0 | 0 | 39 | 275 | 50 | 0.15 | 0.20 | 0.10 | 0.22 | 0.20 | 0.36 | 1.08 | 5.64 | 16.13 | 21.77 |
| 15 | −1.68 | −1.68 | −1.68 | 1 | 50 | 0 | 0.04 | 0.03 | 0.03 | 0.04 | 0.04 | 0.02 | 0.16 | 0.95 | 2.46 | 3.41 |
| 16 | 1.68 | −1.68 | −1.68 | 39 | 50 | 0 | 0.06 | 0.05 | 0.04 | 0.05 | 0.06 | 0.06 | 0.26 | 1.14 | 3.09 | 4.23 |
| 17 | −1.68 | 1.68 | −1.68 | 1 | 500 | 0 | 0.04 | 0.04 | 0.03 | 0.04 | 0.04 | 0.05 | 0.21 | 1.00 | 2.53 | 3.53 |
| 18 | 1.68 | 1.68 | −1.68 | 39 | 500 | 0 | 0.13 | 0.11 | 0.09 | 0.11 | 0.12 | 0.14 | 0.57 | 1.14 | 4.74 | 5.88 |
| 19 | −1.68 | −1.68 | 1.68 | 1 | 50 | 100 | 0.08 | 0.12 | 0.06 | 0.14 | 0.11 | 0.17 | 0.60 | 5.22 | 9.65 | 14.87 |
| 20 | 1.68 | −1.68 | 1.68 | 39 | 50 | 100 | 0.11 | 0.15 | 0.09 | 0.19 | 0.15 | 0.24 | 0.82 | 5.58 | 13.42 | 19.00 |
| 21 | −1.68 | 1.68 | 1.68 | 1 | 500 | 100 | 0.08 | 0.12 | 0.04 | 0.12 | 0.10 | 0.15 | 0.52 | 4.26 | 10.50 | 14.76 |
| 22 | 1.68 | 1.68 | 1.68 | 39 | 500 | 100 | 0.13 | 0.07 | 0.04 | 0.08 | 0.07 | 0.01 | 0.27 | 0.97 | 7.05 | 8.02 |
| 23 | 0 | 0 | 0 | 20 | 275 | 50 | 0.14 | 0.20 | 0.08 | 0.22 | 0.24 | 0.35 | 1.09 | 5.46 | 15.56 | 21.02 |
| 24 | 0 | 0 | 0 | 20 | 275 | 50 | 0.15 | 0.20 | 0.10 | 0.23 | 0.22 | 0.36 | 1.10 | 4.25 | 16.80 | 21.05 |
| 25 | 0 | 0 | 0 | 20 | 275 | 50 | 0.12 | 0.25 | 0.10 | 0.25 | 0.22 | 0.35 | 1.16 | 5.08 | 16.35 | 21.43 |
| 26 | 0 | 0 | 0 | 20 | 275 | 50 | 0.14 | 0.21 | 0.10 | 0.25 | 0.23 | 0.30 | 1.11 | 4.34 | 15.46 | 19.81 |
| 27 | 0 | 0 | 0 | 20 | 275 | 50 | 0.14 | 0.25 | 0.09 | 0.23 | 0.22 | 0.25 | 1.03 | 4.68 | 16.10 | 20.78 |
| 28 | 0 | 0 | 0 | 20 | 275 | 50 | 0.14 | 0.26 | 0.14 | 0.23 | 0.24 | 0.28 | 1.15 | 4.96 | 15.47 | 20.43 |
| PARAMETERS | RESIDUE | FLAVONOID CONTENT | TANNIN CONTENT | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| INDIVIDUAL | TOTAL | INDIVIDUAL | TOTAL | ||||||||||||||||||
| Yield | F1 | F2 | F3 | F4 | F5 | TF | T1 | T2 | TT | ||||||||||||
| Intercept | b0 | 0.140 | ±0.004 | 0.224 | ±0.011 | 0.100 | ±0.006 | 0.233 | ±0.010 | 0.227 | ±0.012 | 0.289 | ±0.016 | 1.106 | ±0.036 | 4.923 | ±0.273 | 15.786 | ±1.306 | 20.312 | ±0.861 |
| Linear Effect | b1 | 0.010 | ±0.002 | −0.017 | ±0.017 | ns | ns | −0.016 | ±0.016 | −0.045 | ±0.032 | −0.083 | ±0.049 | ns | 2.061 | ±0.531 | 2.744 | ±0.452 | |||
| b2 | 0.026 | ±0.007 | ns | ns | ns | ns | ns | ns | ns | ns | ns | ||||||||||
| b3 | −0.011 | ±0.007 | −0.018 | ±0.017 | 0.037 | ±0.012 | 0.006 | ±0.004 | ns | 0.009 | ±0.009 | 0.043 | ±0.015 | −0.207 | ±0.164 | ns | −3.754 | ±1.515 | |||
| Quadratic Effect | b11 | −0.014 | ±0.003 | −0.045 | ±0.009 | −0.006 | ±0.006 | −0.051 | ±0.007 | −0.046 | ±0.008 | −0.064 | ±0.009 | −0.206 | ±0.026 | −0.827 | ±0.154 | −3.077 | ±0.864 | −3.628 | ±0.745 |
| b22 | ns | ns | ns | −0.005 | ±0.007 | −0.018 | ±0.008 | ns | −0.045 | ±0.026 | ns | ns | ns | ||||||||
| b33 | −0.005 | ±0.003 | −0.020 | ±0.009 | ns | −0.011 | ±0.007 | −0.024 | ±0.008 | ns | −0.061 | ±0.026 | ns | −0.812 | ±0.864 | ns | |||||
| Cubic Effect | b111 | ns | 0.012 | ±0.007 | ns | 0.008 | ±0.002 | 0.010 | ±0.006 | 0.023 | ±0.013 | 0.055 | ±0.019 | 0.294 | ±0.064 | ns | ns | ||||
| b222 | −0.006 | ±0.003 | ns | ns | ns | ns | ns | ns | ns | ns | ns | ||||||||||
| b333 | 0.009 | ±0.003 | 0.009 | ±0.007 | −0.009 | ±0.005 | ns | ns | ns | ns | ns | ns | 1.462 | ±0.596 | |||||||
| Interactive Linear Effect | b12 | ns | −0.005 | ±0.004 | −0.005 | ±0.003 | −0.010 | ±0.003 | −0.007 | ±0.003 | −0.016 | ±0.007 | −0.043 | ±0.010 | −0.260 | ±0.116 | ns | −0.498 | ±0.321 | ||
| b13 | ns | −0.004 | ±0.004 | ns | ns | −0.003 | ±0.003 | ns | −0.015 | ±0.010 | ns | ns | ns | ||||||||
| b23 | 0.003 | ±0.002 | ns | ns | ns | ns | −0.007 | ±0.001 | ns | −0.173 | ±0.116 | ns | −0.380 | ±0.321 | |||||||
| b123 | −0.002 | ±0.001 | −0.004 | ±0.002 | −0.002 | ±0.002 | −0.004 | ±0.002 | −0.003 | ±0.002 | −0.006 | ±0.004 | −0.018 | ±0.006 | −0.077 | ±0.072 | ns | −0.307 | ±0.198 | ||
| Interactive Quadratic Effect | b1122 | ns | ns | ns | ns | ns | ns | ns | ns | −4.273 | ±0.864 | −5.336 | ±0.952 | ||||||||
| b1133 | ns | ns | ns | ns | ns | ns | −0.135 | ±0.071 | ns | ns | ns | ||||||||||
| b2233 | ns | −0.069 | ±0.023 | ns | ns | ns | ns | ns | ns | ns | ns | ||||||||||
| b112233 | ns | 0.026 | ±0.008 | −0.001 | ±0.001 | 0.002 | ±0.001 | 0.005 | ±0.002 | ns | 0.057 | ±0.022 | ns | 1.594 | ±0.176 | 1.849 | ±0.657 | ||||
| Statistics (R²) | 0.9331 | 0.9066 | 0.8160 | 0.9408 | 0.9036 | 0.8473 | 0.9641 | 0.8305 | 0.8426 | 0.9206 | |||||||||||
| OPTIMAL VARIABLE CONDITIONS | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| X1: t (min) | X2: P (W) | X3: S (%) | ||||||||
| A) INDIVIDUAL OPTIMAL RESPONSE VALUES | ||||||||||
| Yield | 39.00 | ±3.74 | 446.34 | ±21.13 | 58.08 | ±7.62 | 0.18 | ±0.04 | g R/g DW | |
| FLAVONOID CONTENT | F1 | 17.64 | ±2.59 | 291.45 | ±17.07 | 43.28 | ±6.58 | 0.23 | ±0.13 | mg F1/g R |
| F2 | 33.06 | ±3.46 | 50.00 | ±7.07 | 65.80 | ±8.11 | 0.13 | ±0.04 | mg F2/g R | |
| F3 | 23.37 | ±2.94 | 275.00 | ±16.58 | 50.00 | ±7.07 | 0.23 | ±0.05 | mg F3/g R | |
| F4 | 20.11 | ±2.75 | 279.01 | ±16.70 | 45.20 | ±6.72 | 0.23 | ±0.05 | mg F4/g R | |
| F5 | 9.31 | ±1.96 | 500.00 | ±22.36 | 46.12 | ±6.79 | 0.30 | ±0.05 | mg F5/g R | |
| FT | 23.92 | ±2.97 | 289.36 | ±17.01 | 44.18 | ±6.65 | 1.12 | ±0.08 | mg TF/g R | |
| TANNIN CONTENT | T1 | 1.00 | ±0.24 | 500.00 | ±22.36 | 46.31 | ±6.81 | 5.78 | ±0.39 | mg T1/g R |
| T2 | 20.00 | ±2.74 | 275.00 | ±16.58 | 59.96 | ±7.74 | 16.13 | ±0.90 | mg T2/g R | |
| TT | 8.52 | ±1.89 | 500.00 | ±22.36 | 52.71 | ±7.26 | 23.35 | ±0.47 | mg TT/g R | |
| B) RELATIVE OPTIMAL RESPONSE VALUES FOR FLAVONOID CONTENT | ||||||||||
| Yield | 26.32 | ±3.07 | 285.57 | ±16.90 | 44.80 | ±6.69 | 0.13 | ±0.07 | g R/g DW | |
| FLAVONOID CONTENT | F1 | 0.21 | ±0.06 | mg F1/g R | ||||||
| F2 | 0.12 | ±0.04 | mg F2/g R | |||||||
| F3 | 0.23 | ±0.08 | mg F3/g R | |||||||
| F4 | 0.22 | ±0.07 | mg F4/g R | |||||||
| F5 | 0.30 | ±0.05 | mg F5/g R | |||||||
| TANNIN CONTENT | T1 | 4.79 | ±0.19 | mg T1/g R | ||||||
| T2 | 15.12 | ±1.89 | mg T2/g R | |||||||
| C) RELATIVE OPTIMAL RESPONSE VALUES FOR TANNIN CONTENT | ||||||||||
| Yield | 11.44 | ±2.03 | 500.00 | ±22.39 | 51.22 | ±7.16 | 0.15 | ±0.09 | g R/g DW | |
| FLAVONOID CONTENT | F1 | 0.07 | ±0.01 | mg F1/g R | ||||||
| F2 | 0.07 | ±0.02 | mg F2/g R | |||||||
| F3 | 0.20 | ±0.05 | mg F3/g R | |||||||
| F4 | 0.16 | ±0.03 | mg F4/g R | |||||||
| F5 | 0.29 | ±0.04 | mg F5/g R | |||||||
| TANNIN CONTENT | T1 | 5.35 | ±0.31 | mg T1/g R | ||||||
| T2 | 15.23 | ±0.90 | mg T2/g R | |||||||
| D) GLOBAL OPTIMAL RESPONSE VALUES | ||||||||||
| Yield | 23.47 | ±2.90 | 258.78 | ±16.09 | 50.51 | ±7.11 | 0.13 | ±0.07 | g R/g DW | |
| FLAVONOID CONTENT | F1 | 0.22 | ±0.07 | mg F1/g R | ||||||
| F2 | 0.11 | ±0.03 | mg F2/g R | |||||||
| F3 | 0.23 | ±0.08 | mg F3/g R | |||||||
| F4 | 0.23 | ±0.07 | mg F4/g R | |||||||
| F5 | 0.29 | ±0.04 | mg F5/g R | |||||||
| TANNIN CONTENT | T1 | 4.88 | ±0.21 | mg T1/g R | ||||||
| T2 | 15.77 | ±0.97 | mg T2/g R | |||||||
| Optimized Extract | Positive Control | ||||||
|---|---|---|---|---|---|---|---|
| Antioxidant Activity (IC50 Values, µg/mL) | Trolox | ||||||
| TBARS Inhibition | 2.7 ± 0.1 | 5.8 ± 0.6 | |||||
| OxHLIA | Δt 60 min | 29 ± 1 | 19.6 ± 0.7 | ||||
| Δt 80 min | 54 ± 2 | 26.9 ± 0.8 | |||||
| Anti-Inflammatory Activity (EC50 Values, µg/mL) | Dexamethasone | ||||||
| RAW 267.4 | 195 ± 8 | 16 ± 1 | |||||
| Cytotoxicity (GI50 Values, µg/mL) | Ellipticine | ||||||
| AGS (gastric carcinoma) | 58 ± 3 | 0.9 ± 0.1 | |||||
| CaCo (colorectal adenocarcinoma) | 39 ± 1 | 0.8 ± 0.1 | |||||
| MCF-7 (breast carcinoma) | 141 ± 5 | 1.020 ± 0.004 | |||||
| NCI-H460 (non-small cell lung cancer) | 232 ± 6 | 1.01 ± 0.01 | |||||
| Hepatotoxicity (GI50 Values, µg/mL) | Dexamethasone | ||||||
| VERO | 110 ± 5 | 0.6 ± 0.1 | |||||
| Antibacterial Activity (mg/mL) | E221 | E224 | |||||
| MIC | MBC | MIC | MBC | MIC | MBC | ||
| Staphylococcus aureus | 1 | 2 | 4 | 4 | 1 | 1 | |
| Bacillus cereus | 0.5 | 1 | 0.5 | 0.5 | 2 | 4 | |
| Listeria monocytogenes | 1 | 2 | 1 | 2 | 0.5 | 1 | |
| Escherichia coli | 0.5 | 1 | 1 | 2 | 0.5 | 1 | |
| Salmonella Typhimurium | 0.5 | 1 | 1 | 2 | 1 | 1 | |
| Enterobacter cloacae | 1 | 2 | 2 | 4 | 0.5 | 0.5 | |
| Antifungal Activity (mg/mL) | E221 | E224 | |||||
| MIC | MFC | MIC | MFC | MIC | MFC | ||
| Aspergillus fumigatus | 2 | 4 | 1 | 2 | 1 | 1 | |
| Aspergillus niger | 1 | 2 | 1 | 2 | 1 | 1 | |
| Aspergillus versicolor | 1 | 2 | 2 | 2 | 1 | 1 | |
| Penicillium funiculosum | 0.25 | 0.5 | 1 | 2 | 0.5 | 0.5 | |
| Penicillium verrucosum var. cyclopium | 1 | 2 | 2 | 4 | 1 | 2 | |
| Trichoderma viride | 0.25 | 0.5 | 1 | 2 | 0.5 | 0.5 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alaya, I.b.; Pereira, E.; Dias, M.I.; Pinela, J.; Calhelha, R.C.; Soković, M.; Kostić, M.; Prieto, M.A.; Essid, F.; Caleja, C.; et al. Development of a Natural Preservative from Chestnut Flowers: Ultrasound-Assisted Extraction Optimization and Functionality Assessment. Chemosensors 2021, 9, 141. https://0-doi-org.brum.beds.ac.uk/10.3390/chemosensors9060141
Alaya Ib, Pereira E, Dias MI, Pinela J, Calhelha RC, Soković M, Kostić M, Prieto MA, Essid F, Caleja C, et al. Development of a Natural Preservative from Chestnut Flowers: Ultrasound-Assisted Extraction Optimization and Functionality Assessment. Chemosensors. 2021; 9(6):141. https://0-doi-org.brum.beds.ac.uk/10.3390/chemosensors9060141
Chicago/Turabian StyleAlaya, Ikbel ben, Eliana Pereira, Maria Inês Dias, José Pinela, Ricardo C. Calhelha, Marina Soković, Marina Kostić, Miguel A. Prieto, Faten Essid, Cristina Caleja, and et al. 2021. "Development of a Natural Preservative from Chestnut Flowers: Ultrasound-Assisted Extraction Optimization and Functionality Assessment" Chemosensors 9, no. 6: 141. https://0-doi-org.brum.beds.ac.uk/10.3390/chemosensors9060141











