BDNF rs6265 Polymorphism and Its Methylation in Patients with Stroke Undergoing Rehabilitation
Abstract
:1. Introduction
2. Results
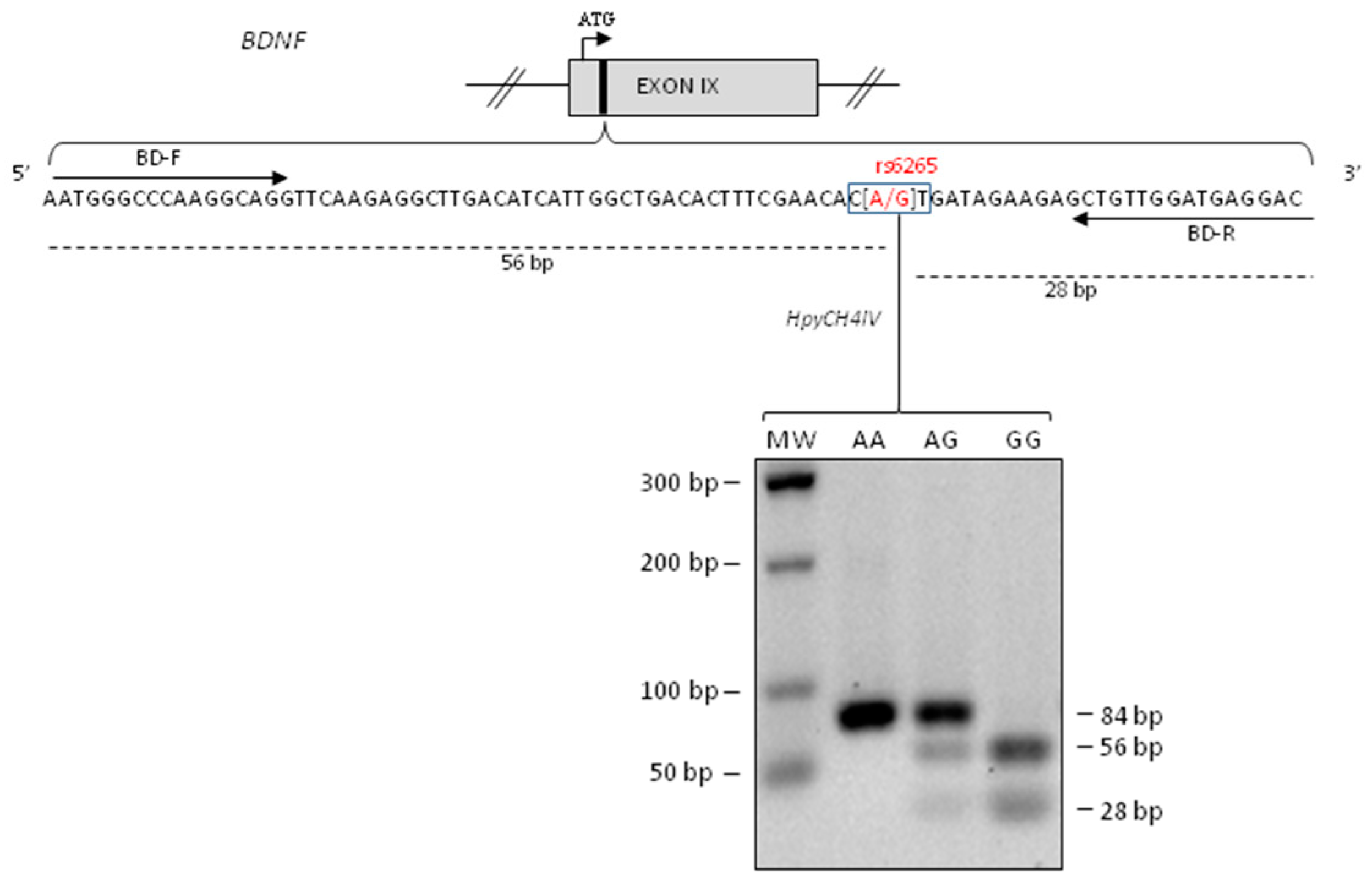
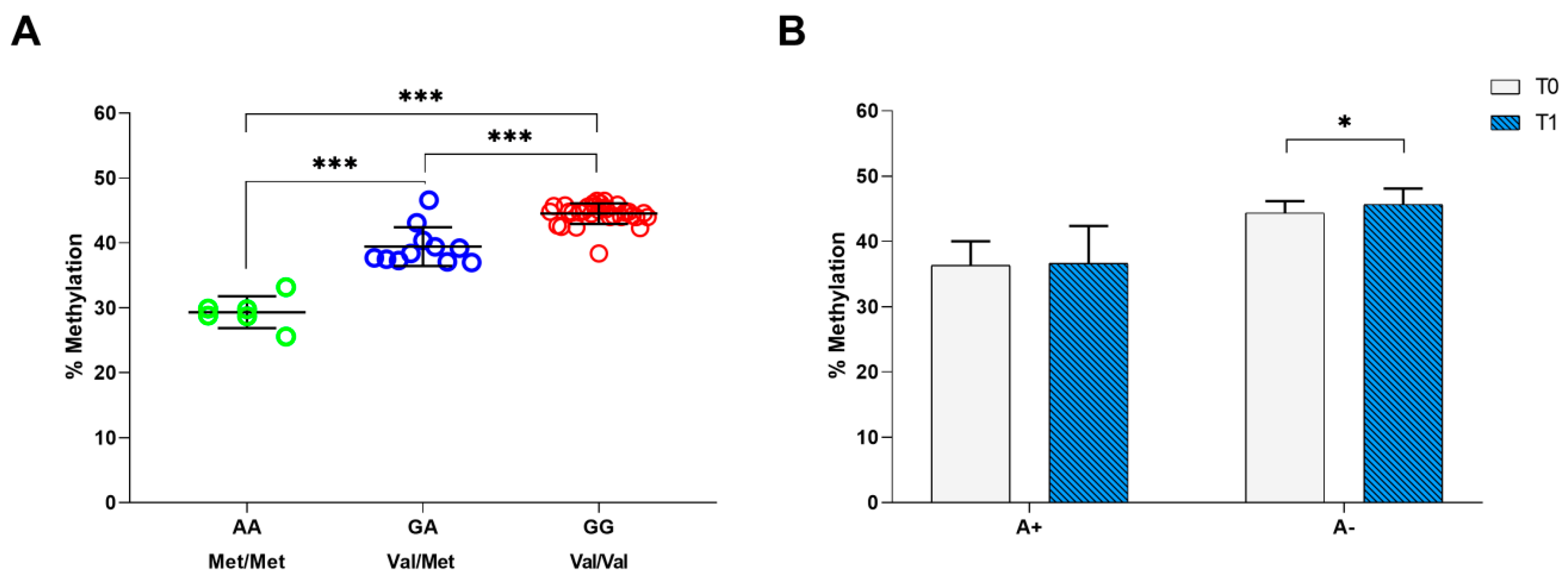
2.1. BDNF rs6265genotyping and Methylation Analysis
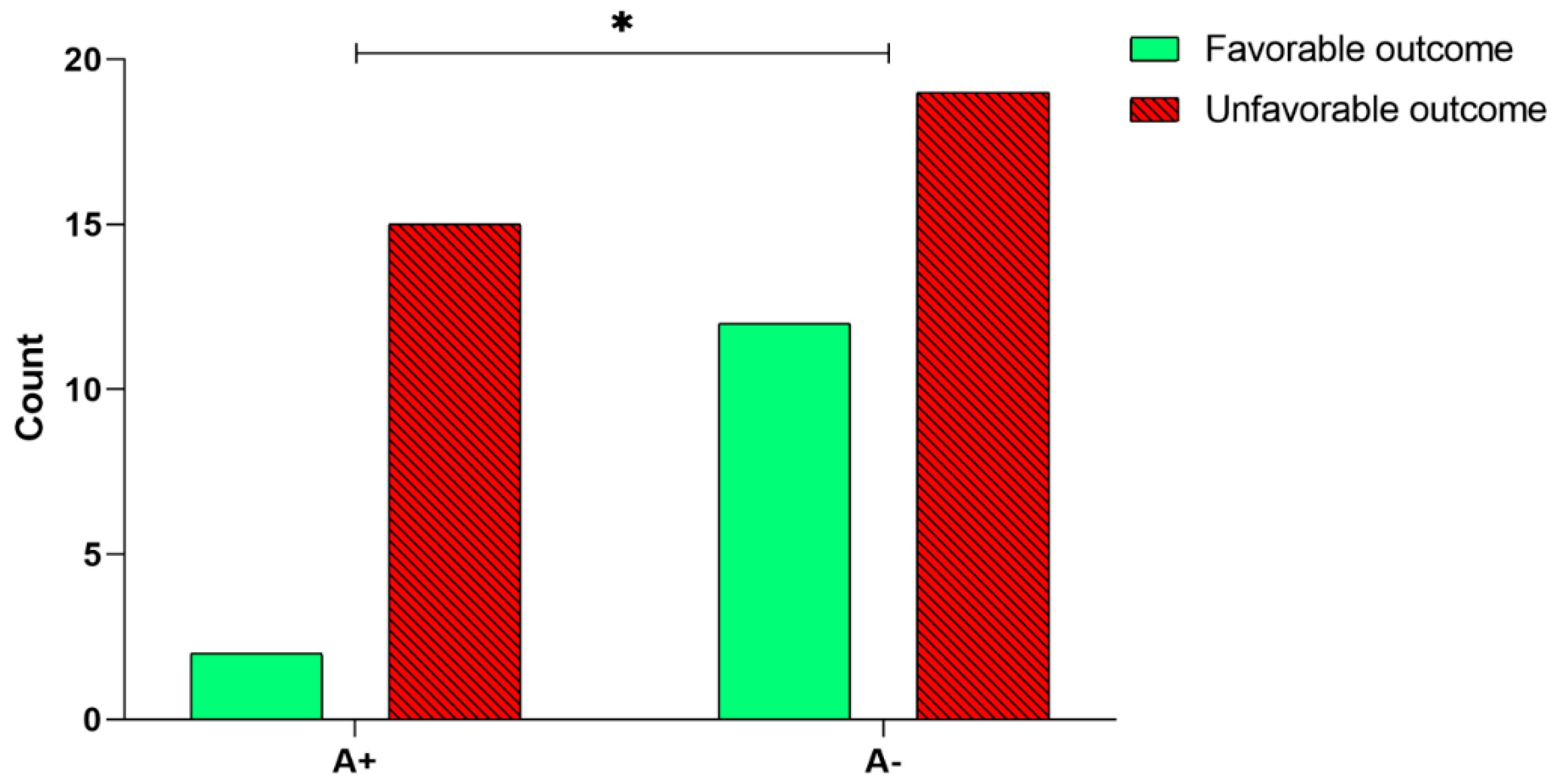
2.2. Relationship between BDNF rs6265 SNP Analysis/Methylation and Rehabilitation Outcome
3. Discussion
4. Materials and Methods
4.1. Sample
4.2. Rehabilitation Treatment and Outcome
4.3. DNA Extraction
4.4. BDNF rs6265 Polymorphism Genotyping
4.5. Bisulphite Conversion
4.6. Methylation Analysis of BDNF rs6265polymorphism
4.7. Statistics
Author Contributions
Funding
Conflicts of Interest
References
- Go, A.S.; Mozaffarian, D.; Roger, V.L.; Benjamin, E.J.; Berry, J.D.; Blaha, M.J.; Dai, S.; Ford, E.S.; Fox, C.S.; Franco, S.; et al. Executive Summary: Heart Disease and Stroke Statistics—2014 Update. Circulation 2014, 129, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Feigin, V.L.; Forouzanfar, M.H.; Krishnamurthi, R.; Mensah, G.A.; Connor, M.; Bennett, D.A.; Moran, A.E.; Sacco, R.L.; Anderson, L.; Truelsen, T.; et al. Global and regional burden of stroke during 1990-2010: Findings from the Global Burden of Disease Study 2010. Lancet 2014, 383, 245–255. [Google Scholar] [CrossRef]
- Prabhakaran, S.; Zarahn, E.; Riley, C.; Speizer, A.; Chong, J.Y.; Lazar, R.M.; Marshall, R.S.; Krakauer, J.W. Inter-individual Variability in the Capacity for Motor Recovery After Ischemic Stroke. Neurorehabil. Neural Repair 2008, 22, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Kwakkel, G.; Wagenaar, R.C.; Kollen, B.J.; Lankhorst, G.J. Predicting Disability in Stroke—A Critical Review of the Literature. Age Ageing 1996, 25, 479–489. [Google Scholar] [CrossRef] [Green Version]
- Vogt, G.; Laage, R.; Shuaib, A.; Schneider, A. Initial Lesion Volume Is an Independent Predictor of Clinical Stroke Outcome at Day 90. Stroke 2012, 43, 1266–1272. [Google Scholar] [CrossRef] [Green Version]
- Saver, J.L.; Johnston, K.C.; Homer, D.; Wityk, R.; Koroshetz, W.; Truskowski, L.L.; Haley, E.C. Infarct volume as a surrogate or auxiliary outcome measure in ischemic stroke clinical trials. The RANTTAS Investigators. Stroke 1999, 30, 293–298. [Google Scholar] [CrossRef]
- Shelton, F.D.N.; Reding, M.J. Effect of Lesion Location on Upper Limb Motor Recovery After Stroke. Stroke 2001, 32, 107–112. [Google Scholar] [CrossRef]
- Cheng, B.; Forkert, N.D.; Zavaglia, M.; Hilgetag, C.C.; Golsari, A.; Siemonsen, S.; Fiehler, J.; Pedraza, S.; Puig, J.; Cho, T.-H.; et al. Influence of Stroke Infarct Location on Functional Outcome Measured by the Modified Rankin Scale. Stroke 2014, 45, 1695–1702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stinear, C.M.; Barber, P.A.; Smale, P.R.; Coxon, J.P.; Fleming, M.K.; Byblow, W.D. Functional potential in chronic stroke patients depends on corticospinal tract integrity. Brain 2006, 130, 170–180. [Google Scholar] [CrossRef]
- Lindenberg, R.; Renga, V.; Zhu, L.L.; Betzler, F.; Alsop, D.; Schlaug, G. Structural integrity of corticospinal motor fibers predicts motor impairment in chronic stroke. Neurology 2010, 74, 280–287. [Google Scholar] [CrossRef] [Green Version]
- Kwakkel, G.; Kollen, B. Predicting improvement in the upper paretic limb after stroke: A longitudinal prospective study. Restor. Neurol. Neurosci. 2007, 25, 453–460. [Google Scholar] [PubMed]
- Stinear, C.M.; Barber, P.A.; Petoe, M.; Anwar, S.; Byblow, W.D. The PREP algorithm predicts potential for upper limb recovery after stroke. Brain 2012, 135, 2527–2535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.Y.; Winstein, C.J. A systematic review of voluntary arm recovery in hemiparetic stroke: Critical predictors for meaningful outcomes using the international classification of functioning, disability, and health. J. Neurol. Phys. Ther. 2009, 33, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, E.B.; Dodakian, L.; See, J.; McKenzie, A.; Le, V.; Wojnowicz, M.; Shahbaba, B.; Cramer, S.C. Neural function, injury, and stroke subtype predict treatment gains after stroke. Ann. Neurol. 2015, 77, 132–145. [Google Scholar] [CrossRef] [Green Version]
- Boyd, L.A.; Hayward, K.S.; Ward, N.S.; Stinear, C.M.; Rosso, C.; Fisher, R.J.; Carter, A.R.; Leff, A.P.; Copland, D.A.; Carey, L.M.; et al. Biomarkers of Stroke Recovery: Consensus-Based Core Recommendations from the Stroke Recovery and Rehabilitation Roundtable. Int. J. Stroke 2017, 12, 480–493. [Google Scholar] [CrossRef]
- Lindgren, A. Stroke Genetics: A Review and Update. J. Stroke 2014, 16, 114. [Google Scholar] [CrossRef]
- Pearson-Fuhrhop, K.M.; Cramer, S.C. Genetic Influences on Neural Plasticity. PM&R 2010, 2, S227–S240. [Google Scholar]
- Cowansage, K.; LeDoux, J.; Monfils, M.-H. Brain-Derived Neurotrophic Factor: A Dynamic Gatekeeper of Neural Plasticity. Curr. Mol. Pharmacol. 2010, 3, 12–29. [Google Scholar] [CrossRef]
- Huang, E.J.; Reichardt, L.F. Neurotrophins: Roles in Neuronal Development and Function. Annu. Rev. Neurosci. 2001, 24, 677–736. [Google Scholar] [CrossRef] [Green Version]
- Lu, B. BDNF and Activity-Dependent Synaptic Modulation. Learn. Mem. 2003, 10, 86–98. [Google Scholar] [CrossRef] [Green Version]
- López-Valdés, H.E.; Clarkson, A.N.; Ao, Y.; Charles, A.C.; Carmichael, S.T.; Sofroniew, M.V.; Brennan, K.C. Memantine enhances recovery from stroke. Stroke 2014, 45, 2093–2100. [Google Scholar] [CrossRef]
- Berretta, A.; Tzeng, Y.-C.; Clarkson, A.N. Post-stroke recovery: The role of activity-dependent release of brain-derived neurotrophic factor. Expert Rev. Neurother. 2014, 14, 1335–1344. [Google Scholar] [CrossRef]
- Clarkson, A.N.; Carmichael, S.T. Cortical excitability and post-stroke recovery. Biochem. Soc. Trans. 2009, 37, 1412–1414. [Google Scholar] [CrossRef] [PubMed]
- Fritsch, B.; Reis, J.; Martinowich, K.; Schambra, H.M.; Ji, Y.; Cohen, L.G.; Lu, B. Direct Current Stimulation Promotes BDNF-Dependent Synaptic Plasticity: Potential Implications for Motor Learning. Neuron 2010, 66, 198–204. [Google Scholar] [CrossRef] [Green Version]
- French, M.A.; Morton, S.M.; Pohlig, R.T.; Reisman, D.S. The relationship between BDNF Val66Met polymorphism and functional mobility in chronic stroke survivors. Top. Stroke Rehabil. 2018, 25, 276–280. [Google Scholar] [CrossRef] [PubMed]
- Shiner, C.T.; Pierce, K.D.; Thompson-Butel, A.G.; Trinh, T.; Schofield, P.R.; McNulty, P.A. BDNF Genotype Interacts with Motor Function to Influence Rehabilitation Responsiveness Poststroke. Front. Neurol. 2016, 7, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.-M.; Stewart, R.; Park, M.-S.; Kang, H.-J.; Kim, S.-W.; Shin, I.-S.; Kim, H.-R.; Shin, M.-G.; Cho, K.-H.; Yoon, J.-S. Associations of BDNF Genotype and Promoter Methylation with Acute and Long-Term Stroke Outcomes in an East Asian Cohort. PLoS ONE 2012, 7, e51280. [Google Scholar] [CrossRef]
- Shimizu, E.; Hashimoto, K.; Iyo, M. Ethnic difference of the BDNF 196G/A (val66met) polymorphism frequencies: The possibility to explain ethnic mental traits. Am. J. Med. Genet. 2004, 126B, 122–123. [Google Scholar] [CrossRef]
- Egan, M.F.; Kojima, M.; Callicott, J.H.; Goldberg, T.E.; Kolachana, B.S.; Bertolino, A.; Zaitsev, E.; Gold, B.; Goldman, D.; Dean, M.; et al. The BDNF val66met Polymorphism Affects Activity-Dependent Secretion of BDNF and Human Memory and Hippocampal Function. Cell 2003, 112, 257–269. [Google Scholar] [CrossRef] [Green Version]
- Kleim, J.A.; Chan, S.; Pringle, E.; Schallert, K.; Procaccio, V.; Jimenez, R.; Cramer, S.C. BDNF val66met polymorphism is associated with modified experience-dependent plasticity in human motor cortex. Nat. Neurosci. 2006, 9, 735–737. [Google Scholar] [CrossRef] [Green Version]
- McHughen, S.A.; Rodriguez, P.F.; Kleim, J.A.; Kleim, E.D.; Crespo, L.M.; Procaccio, V.; Cramer, S.C. BDNF Val66Met Polymorphism Influences Motor System Function in the Human Brain. Cereb. Cortex 2010, 20, 1254–1262. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Kim, E.; Ratan, R.; Lee, F.S.; Cho, S. Genetic Variant of BDNF (Val66Met) Polymorphism Attenuates Stroke-Induced Angiogenic Responses by Enhancing Anti-Angiogenic Mediator CD36 Expression. J. Neurosci. 2011, 31, 775–783. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.-M.; Stewart, R.; Kang, H.-J.; Kim, S.-Y.; Kim, S.-W.; Shin, I.-S.; Park, M.-S.; Kim, H.-R.; Shin, M.-G.; Cho, K.-H.; et al. A longitudinal study of BDNF promoter methylation and genotype with poststroke depression. J. Affect. Disord. 2013, 149, 93–99. [Google Scholar] [CrossRef]
- Martinowich, K.; Hattori, D.; Wu, H.; Fouse, S.; He, F.; Hu, Y.; Fan, G.; Sun, Y.E. DNA Methylation-Related Chromatin Remodeling in Activity-Dependent Bdnf Gene Regulation. Science 2003, 302, 890–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mill, J.; Tang, T.; Kaminsky, Z.; Khare, T.; Yazdanpanah, S.; Bouchard, L.; Jia, P.; Assadzadeh, A.; Flanagan, J.; Schumacher, A.; et al. Epigenomic Profiling Reveals DNA-Methylation Changes Associated with Major Psychosis. Am. J. Hum. Genet. 2008, 82, 696–711. [Google Scholar] [CrossRef] [Green Version]
- Santoro, M.; Nociti, V.; De Fino, C.; Caprara, A.; Giordano, R.; Palomba, N.P.; Losavio, F.; Marra, C.; Patanella, A.K.; Mirabella, M.; et al. Depression in multiple sclerosis: Effect of brain derived neurotrophic factor Val66Met polymorphism and disease perception. Eur. J. Neurol. 2016, 23, 630–640. [Google Scholar] [CrossRef] [PubMed]
- Béjot, Y.; Mossiat, C.; Giroud, M.; Prigent-Tessier, A.; Marie, C. Circulating and brain BDNF levels in stroke rats. relevance to clinical studies. PLoS ONE 2011, 6, e29405. [Google Scholar] [CrossRef] [PubMed]
- Murphy, T.H.; Corbett, D. Plasticity during stroke recovery: From synapse to behaviour. Nat. Rev. Neurosci. 2009, 10, 861–872. [Google Scholar] [CrossRef]
- Chen, Z.Y.; Ieraci, A.; Teng, H.; Dall, H.; Meng, C.X.; Herrera, D.G.; Nykjaer, A.; Hempstead, B.L.; Lee, F.S. Sortilin controls intracellular sorting of brain-derived neurotrophic factor to the regulated secretory pathway. J. Neurosci. 2005, 25, 6156–6166. [Google Scholar] [CrossRef]
- Bergman, Y.; Cedar, H. DNA methylation dynamics in health and disease. Nat. Struct. Mol. Biol. 2013, 20, 274–281. [Google Scholar] [CrossRef]
- Ursini, G.; Cavalleri, T.; Fazio, L.; Angrisano, T.; Iacovelli, L.; Porcelli, A.; Maddalena, G.; Punzi, G.; Mancini, M.; Gelao, B.; et al. BDNF rs6265 methylation and genotype interact on risk for schizophrenia. Epigenetics 2016, 11, 11–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, S.; Vanclay, F.; Cooper, B. Improving the sensitivity of the Barthel Index for stroke rehabilitation. J. Clin. Epidemiol. 1989, 42, 703–709. [Google Scholar] [CrossRef]
- Nociti, V.; Santoro, M.; Quaranta, D.; Losavio, F.A.; De Fino, C.; Giordano, R.; Palomba, N.; Rossini, P.M.; Guerini, F.R.; Clerici, M.; et al. BDNF rs6265 polymorphism methylation in Multiple Sclerosis: A possible marker of disease progression. PLoS ONE 2018, 13, e0206140. [Google Scholar] [CrossRef] [PubMed]
- Eads, C.A.; Danenberg, K.D.; Kawakami, K.; Saltz, L.B.; Blake, C.; Shibata, D.; Danenberg, P.V.; Laird, P.W. MethyLight: A high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000, 28, e32-00. [Google Scholar] [CrossRef] [Green Version]
- Cottrell, S.; Jung, K.; Kristiansen, G.; Eltze, E.; Semjonow, A.; Ittmann, M.; Hartmann, A.; Stamey, T.; Haefliger, C.; Weiss, G. Discovery and Validation of 3 Novel DNA Methylation Markers of Prostate Cancer Prognosis. J. Urol. 2007, 177, 1753–1758. [Google Scholar] [CrossRef]
- Uyttenboogaart, M.; Stewart, R.E.; Vroomen, P.C.A.J.; De Keyser, J.; Luijckx, G.J. Optimizing cutoff scores for the Barthel Index and the modified Rankin Scale for defining outcome in acute stroke trials. Stroke 2005, 36, 1984–1987. [Google Scholar] [CrossRef] [Green Version]
| Variable | Mean (SD), or Count (%) |
|---|---|
| Age | 68.4 (14.3) |
| Sex | 26 men (53.1%) 23 women (46.9%) |
| Time since stroke (days) | 90.2 (31.2) |
| Type of stroke | 36 ischemic (73.5%) 13 hemorrhagic (26.5%) |
| Hemiparesis side | 20 right (40.8%) 29 left (59.2%) |
| Spatial Neglect | 10 (26.3%) |
| Language impairment | 9 (23.7%) |
| Modified Barthel Index | 39.1 (17.2) |
| B | SE | Wald | gl | P | OR | 95% CI OR | |
|---|---|---|---|---|---|---|---|
| Polymorphism * | 1.72 | 0.87 | 3.93 | 1 | 0.047 | 5.59 | 1.02‒30.64 |
| Time Since Stroke | −0.01 | 0.01 | 0.66 | 1 | 0.416 | 0.99 | 0.97‒1.01 |
| Age | −0.03 | 0.02 | 1.31 | 1 | 0.252 | 0.97 | 0.93‒1.02 |
| Constant | 0.53 | 2.02 | 0.07 | 1 | 0.791 | 1.71 |
| Percentage of Methylation | Unfavorable Outcome | Favorable Outcome | p-Value | ||
|---|---|---|---|---|---|
| Mean | SD | Mean | SD | ||
| At Baseline | 41.0 | 5.5 | 42.5 | 5.4 | 0.372 |
| After the treatment | 41.8 | 6.1 | 45.1 | 3.7 | 0.088 |
| Changes from baseline | 0.7 | 2.3 | 1.9 | 2.4 | 0.219 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santoro, M.; Siotto, M.; Germanotta, M.; Bray, E.; Mastrorosa, A.; Galli, C.; Papadopoulou, D.; Aprile, I. BDNF rs6265 Polymorphism and Its Methylation in Patients with Stroke Undergoing Rehabilitation. Int. J. Mol. Sci. 2020, 21, 8438. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21228438
Santoro M, Siotto M, Germanotta M, Bray E, Mastrorosa A, Galli C, Papadopoulou D, Aprile I. BDNF rs6265 Polymorphism and Its Methylation in Patients with Stroke Undergoing Rehabilitation. International Journal of Molecular Sciences. 2020; 21(22):8438. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21228438
Chicago/Turabian StyleSantoro, Massimo, Mariacristina Siotto, Marco Germanotta, Elisa Bray, Alessia Mastrorosa, Camilla Galli, Dionysia Papadopoulou, and Irene Aprile. 2020. "BDNF rs6265 Polymorphism and Its Methylation in Patients with Stroke Undergoing Rehabilitation" International Journal of Molecular Sciences 21, no. 22: 8438. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21228438








