Impact of One-Carbon Metabolism-Driving Epitranscriptome as a Therapeutic Target for Gastrointestinal Cancer
Abstract
:1. Introduction
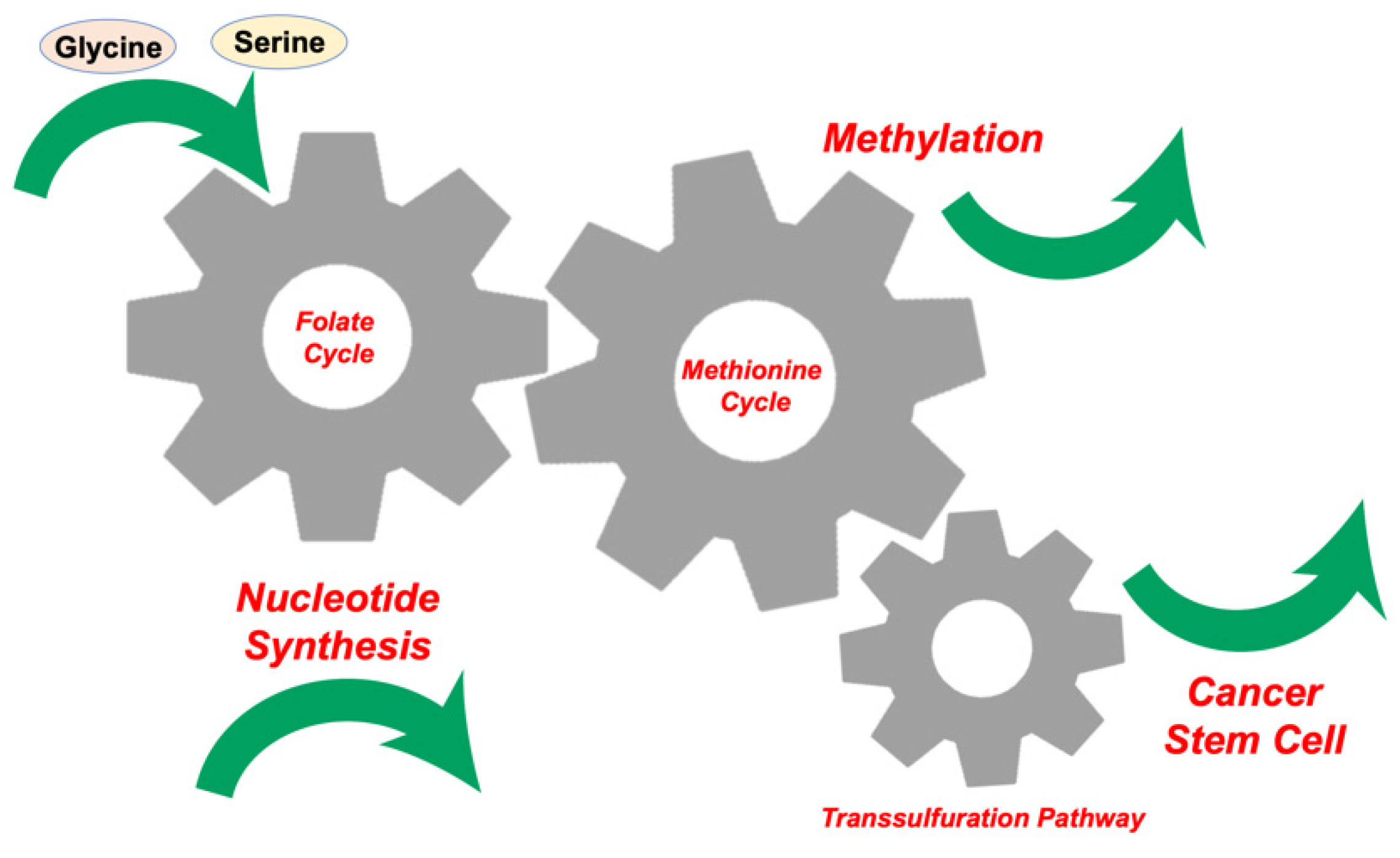
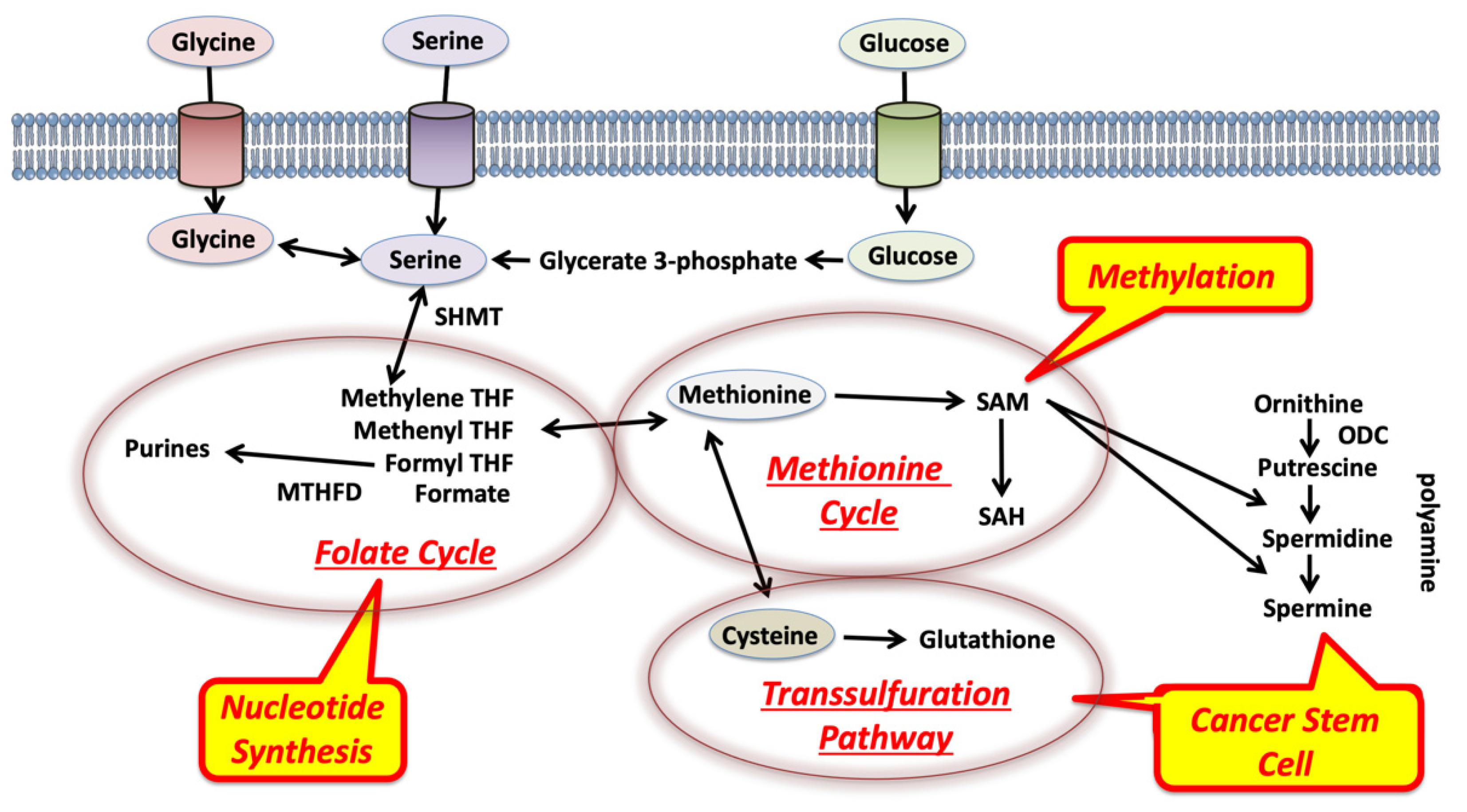
2. Glycine, Serine, and Methionine Control 1C Metabolism in the Tumor Microenvironment
3. Competition for Methionine Upstream in 1C Metabolism
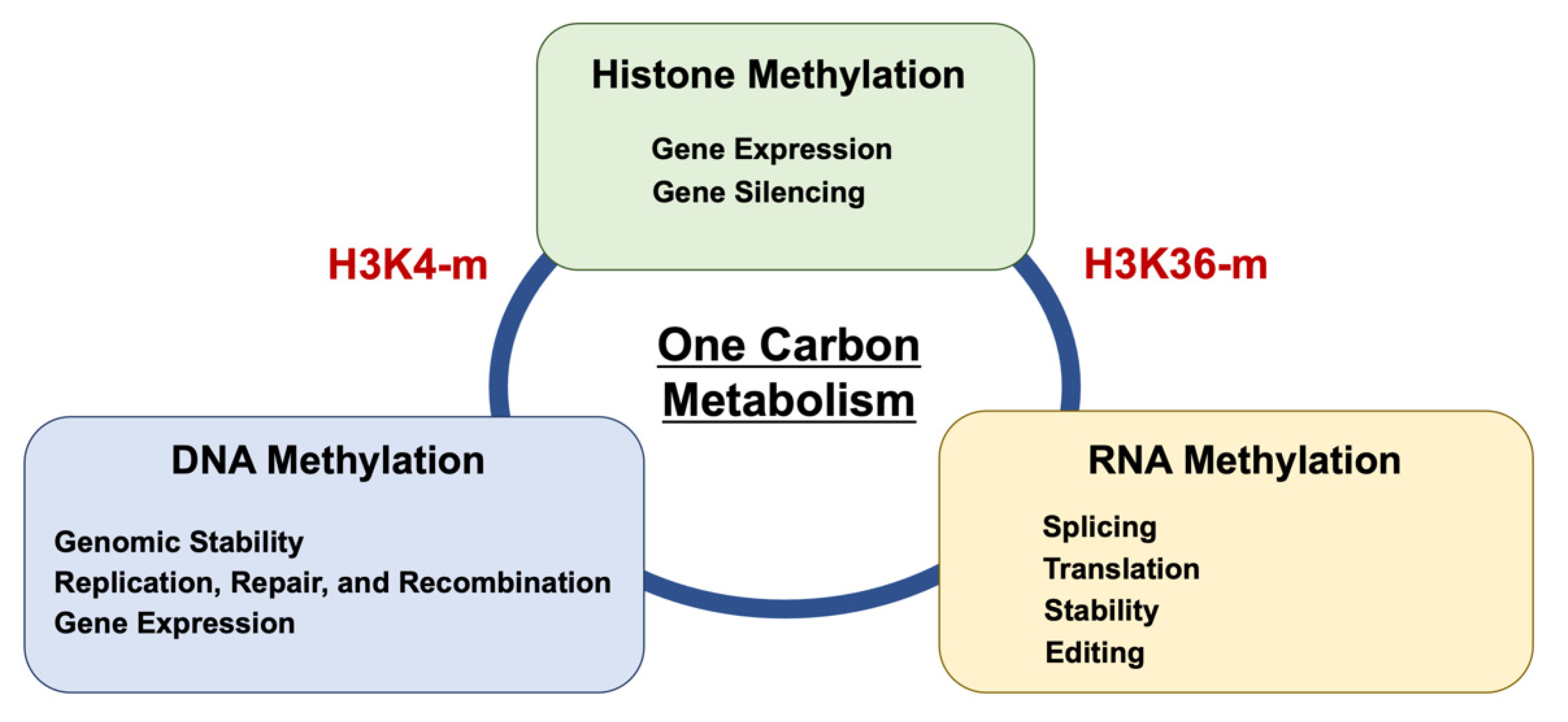
4. Epitranscriptome (RNA Methylation)
5. The Significance of RNA Modification
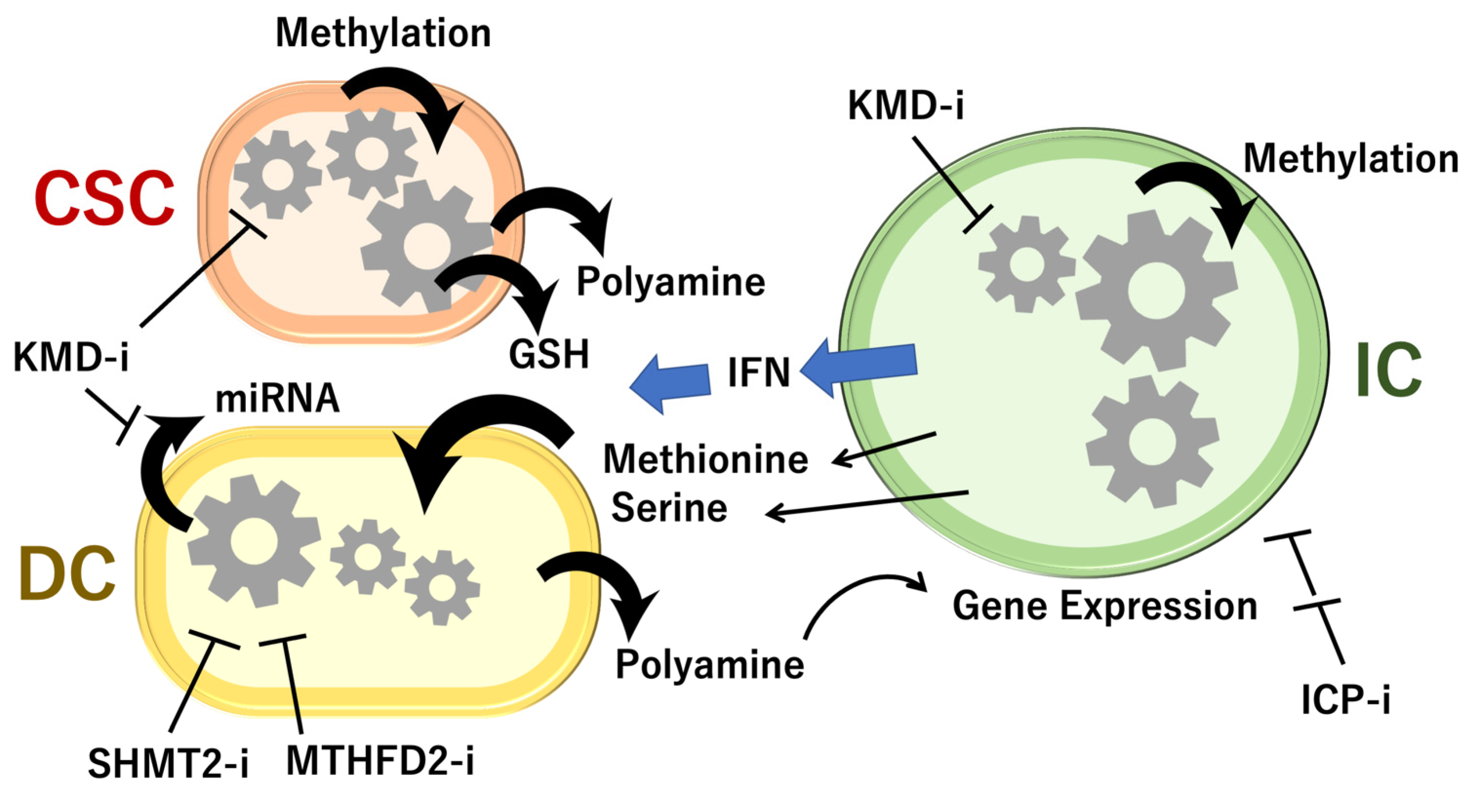
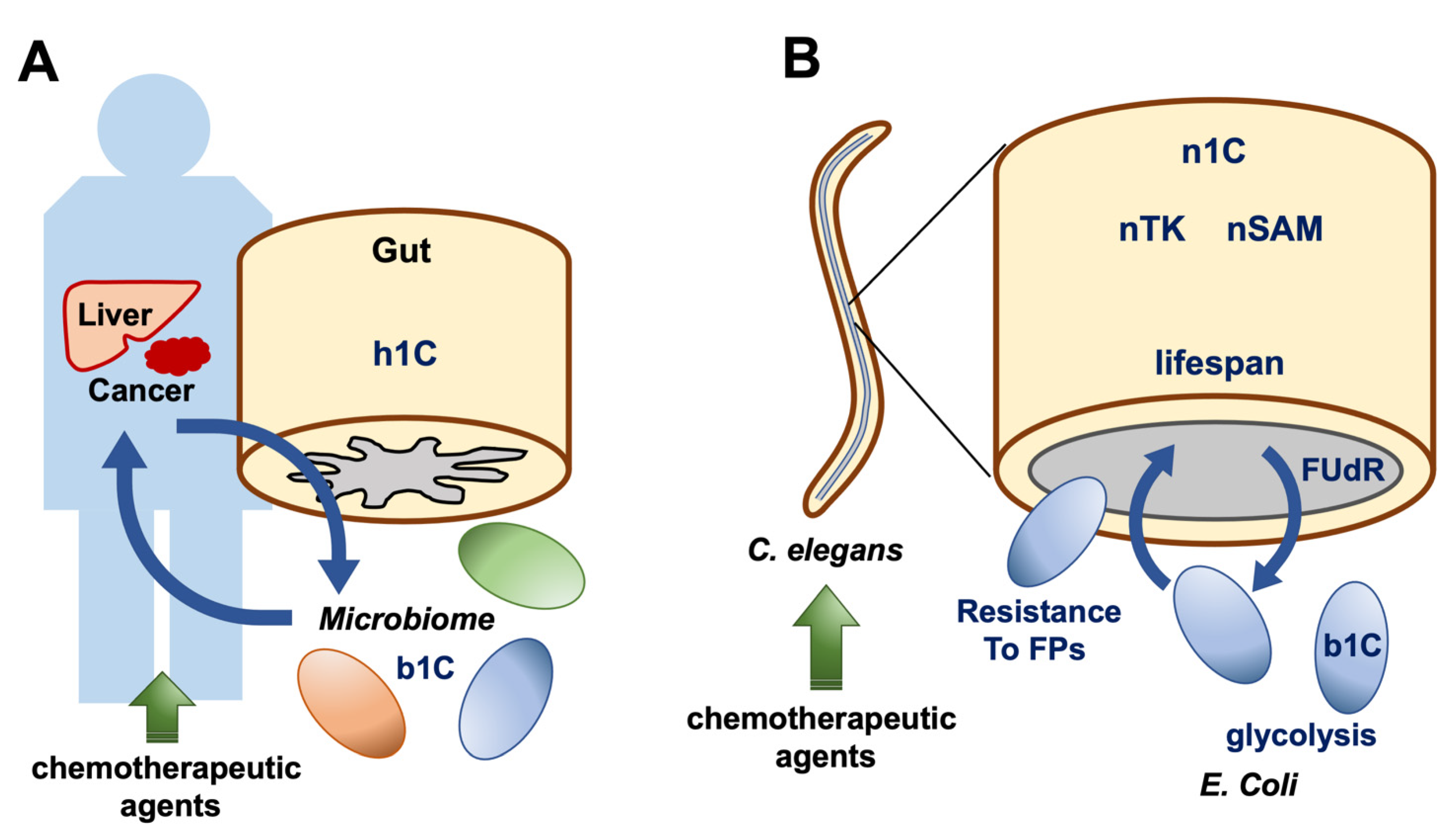
6. Animal Models Reveal Cell-to-Cell Interactions of 1C Metabolites
7. RNA Methylation in Caenorhabditis elegans
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brázda, V.; Bartas, M.; Bowater, R.P. Evolution of Diverse Strategies for Promoter Regulation. Trend. Genet. 2021. [Google Scholar] [CrossRef]
- Jin, W.B.; Wu, S.; Jian, X.H.; Yuan, H.; Tang, G.L. A radical S-adenosyl-L-methionine enzyme and a methyltransferase catalyze cyclopropane formation in natural product biosynthesis. Nat. Commun. 2018, 9, 2771. [Google Scholar] [CrossRef] [Green Version]
- Asai, A.; Konno, M.; Koseki, J.; Taniguchi, M.; Vecchione, A.; Ishii, H. One-carbon metabolism for cancer diagnostic and therapeutic approaches. Cancer Lett. 2020, 470, 141–148. [Google Scholar] [CrossRef]
- Koseki, J.; Konno, M.; Asai, A.; Colvin, H.; Kawamoto, K.; Nishida, N.; Sakai, D.; Kudo, T.; Satoh, T.; Doki, Y.; et al. Enzymes of the one-carbon folate metabolism as anticancer targets predicted by survival rate analysis. Sci. Rep. 2018, 8, 303. [Google Scholar] [CrossRef] [Green Version]
- Konno, M.; Asai, A.; Kawamoto, K.; Nishida, N.; Satoh, T.; Doki, Y.; Mori, M.; Ishii, H. The one-carbon metabolism pathway highlights therapeutic targets for gastrointestinal cancer (Review). Int. J. Oncol. 2017, 50, 1057–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuthbertson, C.R.; Arabzada, Z.; Bankhead, A., 3rd; Kyani, A.; Neamati, N. A review of small-molecule inhibitors of one-carbon enzymes: SHMT2 and MTHFD2 in the spotlight. ACS Pharmacol. Transl. Sci. 2021, 4, 624–646. [Google Scholar] [CrossRef] [PubMed]
- Willbanks, A.; Wood, S.; Cheng, J.X. RNA Epigenetics: Fine-tuning chromatin plasticity and transcriptional regulation, and the implications in human diseases. Genes 2021, 12, 627. [Google Scholar] [CrossRef] [PubMed]
- Konno, M.; Koseki, J.; Asai, A.; Yamagata, A.; Shimamura, T.; Motooka, D.; Okuzaki, D.; Kawamoto, K.; Mizushima, T.; Eguchi, H.; et al. Distinct methylation levels of mature microRNAs in gastrointestinal cancers. Nat. Commun. 2019, 10, 3888. [Google Scholar] [CrossRef] [Green Version]
- Han, Z.; Yang, B.; Wang, Y.; Zeng, X.; Tian, Z. Identification of expression patterns and potential prognostic significance of m5C-related regulators in head and neck squamous cell carcinoma. Front. Oncol. 2021, 11, 612. [Google Scholar] [CrossRef]
- Liu, Y.; Liang, G.; Xu, H.; Dong, W.; Dong, Z.; Qiu, Z.; Zhang, Z.; Li, F.; Huang, Y.; Li, Y.; et al. Tumors exploit FTO-mediated regulation of glycolytic metabolism to evade immune surveillance. Cell Metab. 2021, 33, 1221–1233.e11. [Google Scholar] [CrossRef] [PubMed]
- Yankova, E.; Blackaby, W.; Albertella, M.; Rak, J.; De Braekeleer, E.; Tsagkogeorga, G.; Pilka, E.S.; Aspris, D.; Legatte, D.; Hendrick, A.G.; et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 2021, in press. [Google Scholar] [CrossRef] [PubMed]
- Selberg, D.B.S.; Aatonen, M.; Koivisto, P.; Siltanen, A.; Mervaala, E.; Kankuri, E.; Karelson, M. Discovery of small molecules that activate RNA methylation through cooperative binding to the METTL3-14-WTAP complex active site. Cell Rep. 2019, 26, 3762–3771.e5. [Google Scholar] [CrossRef] [Green Version]
- Cai, Y.; Feng, R.; Lu, T.; Chen, X.; Zhou, X.; Wang, X. Novel insights into the m6A-RNA methyltransferase METTL3 in cancer. Biomark Res. 2021, 9, 27. [Google Scholar] [CrossRef] [PubMed]
- Konno, M.; Taniguchi, M.; Ishii, H. Significant epitranscriptomes in heterogeneous cancer. Cancer Sci. 2019, 110, 2318–2327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bröer, S.; Fairweather, S.J. Amino acid transport across the mammalian intestine. Compr. Physiol. 2018, 9, 343–373. [Google Scholar]
- Locasale, J.W. Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nat. Rev. Cancer 2013, 13, 572–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ducker, G.S.; Rabinowitz, J.D. One-carbon metabolism in health and disease. Cell Metab. 2017, 25, 27–42. [Google Scholar] [CrossRef] [Green Version]
- Cedar, H.; Bergman, Y. Linking DNA methylation and histone modification: Patterns and paradigms. Nat. Rev. Genet. 2009, 10, 295–304. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Zhou, K.; Wu, T.; Zhao, B.S.; Sun, M.; Chen, Z.; Deng, X.; Xiao, G.; Auer, F.; et al. Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally. Nature 2019, 567, 414–419. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, P.; Xia, Q. Epigenetic methylations on N6-adenine and N6-adenosine with the same input but different output. Int. J. Mol. Sci. 2019, 20, 2931. [Google Scholar] [CrossRef] [Green Version]
- Stepka, P.; Vsiansky, V.; Raudenska, M.; Gumulec, J.; Adam, V.; Masarik, M. Metabolic and amino acid alterations of the tumor microenvironment. Curr. Med. Chem. 2021, 28, 1270–1289. [Google Scholar] [CrossRef]
- Cluntun, A.A.; Lukey, M.J.; Cerione, R.A.; Locasale, J.W. Glutamine metabolism in cancer: Understanding the heterogeneity. Trends Cancer 2017, 3, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sreekumar, A.; Poisson, L.M.; Rajendiran, T.M.; Khan, A.P.; Cao, Q.; Yu, J.; Laxman, B.; Mehra, R.; Lonigro, R.J.; Li, Y.; et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 2009, 457, 910–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meléndez-Rodríguez, F.; Urrutia, A.A.; Lorendeau, D.; Rinaldi, G.; Roche, O.; Böğürcü-Seidel, N.; Muelas, M.O.; Mesa-Ciller, C.; Turiel, G.; Bouthelier, A.; et al. HIF1alpha suppresses tumor cell proliferation through inhibition of aspartate biosynthesis. Cell Rep. 2019, 26, 2257–2265.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bansal, A.; Simon, M.C. Glutathione metabolism in cancer progression and treatment resistance. J. Cell Biol. 2018, 217, 2291–2298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Zou, W. Amino acids and their transporters in T cell immunity and cancer therapy. Mol. Cell 2020, 80, 384–395. [Google Scholar] [CrossRef] [PubMed]
- Gubser, P.M.; Kallies, A. Methio "mine"! Cancer cells steal methionine and impair CD8 T-cell function. Immunol. Cell Biol. 2020, 98, 623–625. [Google Scholar] [CrossRef] [PubMed]
- Bian, Y.; Li, W.; Kremer, D.M.; Sajjakulnukit, P.; Li, S.; Crespo, J.; Nwosu, Z.C.; Zhang, L.; Czerwonka, A.; Pawłowska, A.; et al. Cancer SLC43A2 alters T cell methionine metabolism and histone methylation. Nature 2020, 585, 277–282. [Google Scholar] [CrossRef]
- Karikó, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 2005, 23, 165–175. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.G.; Chen, R.; Ahmad, S.; Verma, R.; Kasturi, S.P.; Amaya, L.; Broughton, J.P.; Kim, J.; Cadena, C.; Pulendran, B.; et al. N6-methyladenosine modification controls circular RNA immunity. Mol. Cell 2019, 76, 96–109.e9. [Google Scholar] [CrossRef]
- Chen, G.; Zhao, Q.; Yuan, B.; Wang, B.; Zhang, Y.; Li, Z.; Du, S.; Zeng, Z. ALKBH5-modified HMGB1-STING activation contributes to radiation induced liver disease via innate immune response. Int. J. Radiat. Oncol. Biol. Phys. 2021. [Google Scholar] [CrossRef]
- Asai, A.; Koseki, J.; Konno, M.; Nishimura, T.; Gotoh, N.; Satoh, T.; Doki, Y.; Mori, M.; Ishii, H. Drug discovery of anticancer drugs targeting methylenetetrahydrofolate dehydrogenase 2. Heliyon 2018, 4, e01021. [Google Scholar] [CrossRef] [Green Version]
- Anchisi, S.; Guerra, J.; Garcin, D. RIG-I ATPase activity and discrimination of self-RNA versus non-self-RNA. mBio 2015, 6, e02349. [Google Scholar] [CrossRef]
- Liu, Y.; Olagnier, D.; Lin, R. Host and viral modulation of RIG-I-mediated antiviral immunity. Front. Immunol. 2017, 7, 662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishii, H.; Iwatsuki, M.; Ieta, K.; Ohta, D.; Haraguchi, N.; Mimori, K.; Mori, M. Cancer stem cells and chemoradiation resistance. Cancer Sci. 2008, 99, 1871–1877. [Google Scholar] [CrossRef] [PubMed]
- Bao, S.; Wu, Q.; McLendon, R.E.; Hao, Y.; Shi, Q.; Hjelmeland, A.B.; Dewhirst, M.W.; Bigner, D.D.; Rich, J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006, 444, 756–760. [Google Scholar] [CrossRef] [PubMed]
- Mori, S.; Akita, H.; Kobayashi, S.; Iwagami, Y.; Yamada, D.; Tomimaru, Y.; Noda, T.; Gotoh, K.; Takeda, Y.; Tanemura, M.; et al. Inhibition of c-MET reverses radiation-induced malignant potential in pancreatic cancer. Cancer Lett. 2021, 512, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Pustovalova, M.; Alhaddad, L.; Blokhina, T.; Smetanina, N.; Chigasova, A.; Chuprov-Netochin, R.; Eremin, P.; Gilmutdinova, I.; Osipov, A.N.; Leonov, S. The CD44high subpopulation of multifraction irradiation-surviving NSCLC cells exhibits partial EMT-program activation and DNA damage response depending on their p53 status. Int. J. Mol. Sci. 2021, 22, 2369. [Google Scholar] [CrossRef] [PubMed]
- Shiokawa, D.; Sakai, H.; Ohata, H.; Miyazaki, T.; Kanda, Y.; Sekine, S.; Narushima, D.; Hosokawa, M.; Kato, M.; Suzuki, Y.; et al. Slow-cycling cancer stem cells regulate progression and chemoresistance in colon cancer. Cancer Res. 2020, 80, 4451–4464. [Google Scholar] [CrossRef]
- Choi, J.E.; Sebastian, C.; Ferrer, C.M.; Lewis, C.A.; Sade-Feldman, M.; LaSalle, T.; Gonye, A.; Lopez, B.G.C.; Abdelmoula, W.M.; Regan, M.S.; et al. A unique subset of glycolytic tumour-propagating cells drives squamous cell carcinoma. Nat. Metab. 2021, 3, 182–195. [Google Scholar] [CrossRef]
- Jogo, T.; Oki, E.; Nakanishi, R.; Ando, K.; Nakashima, Y.; Kimura, Y.; Saeki, H.; Oda, Y.; Maehara, Y.; Mori, M. Expression of CD44 variant 9 induces chemoresistance of gastric cancer by controlling intracellular reactive oxygen spices accumulation. Gastric Cancer 2021, in press. [Google Scholar]
- Koseki, J.; Matsui, H.; Konno, M.; Nishida, N.; Kawamoto, K.; Kano, Y.; Mori, M.; Doki, Y.; Ishii, H. A Trans-omics mathematical analysis reveals novel functions of the ornithine metabolic pathway in cancer stem cells. Sci. Rep. 2016, 6, 20726. [Google Scholar] [CrossRef] [Green Version]
- Guengerich, F.P. Introduction: Metals in biology: Alpha-ketoglutarate/iron-dependent dioxygenases. J. Biol. Chem. 2015, 290, 20700–20701. [Google Scholar] [CrossRef] [Green Version]
- Roesch, A.; Fukunaga-Kalabis, M.; Schmidt, E.C.; Zabierowski, S.E.; Brafford, P.A.; Vultur, A.; Basu, D.; Gimotty, P.; Vogt, T.; Herlyn, M. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell 2010, 141, 583–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohta, K.; Haraguchi, N.; Kano, Y.; Kagawa, Y.; Konno, M.; Nishikawa, S.; Hamabe, A.; Hasegawa, S.; Ogawa, H.; Fukusumi, T.; et al. Depletion of Jarid1b induces cellular senescence in human colorectal cancer. Int. J. Oncol. 2013, 42, 1212–1218. [Google Scholar] [CrossRef] [Green Version]
- Dmello, R.S.; To, S.Q.; Chand, A.L. Therapeutic targeting of the tumour microenvironment in metastatic colorectal cancer. Int. J. Mol. Sci. 2021, 22, 2067. [Google Scholar] [CrossRef] [PubMed]
- Taketo, K.; Konno, M.; Asai, A.; Koseki, J.; Toratani, M.; Satoh, T.; Doki, Y.; Mori, M.; Ishii, H.; Ogawa, K. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int. J. Oncol. 2018, 52, 621–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishizawa, Y.; Konno, M.; Asai, A.; Koseki, J.; Kawamoto, K.; Miyoshi, N.; Takahashi, H.; Nishida, N.; Haraguchi, N.; Sakai, D.; et al. Oncogene c-Myc promotes epitranscriptome m6A reader YTHDF1 expression in colorectal cancer. Oncotarget 2017, 9, 7476–7486. [Google Scholar] [CrossRef] [Green Version]
- Jain, M.; Nilsson, R.; Sharma, S.; Madhusudhan, N.; Kitami, T.; Souza, A.L.; Kafri, R.; Kirschner, M.W.; Clish, C.B.; Mootha, V.K. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 2012, 336, 1040–1044. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Torres, I.; Zuniga-Munoz, A.M.; Guarner-Lans, V. Beneficial effects of the amino acid glycine. Mini Rev. Med. Chem. 2017, 17, 15–32. [Google Scholar] [CrossRef]
- Mattaini, K.R.; Sullivan, M.R.; Vander Heiden, M.G. The importance of serine metabolism in cancer. J. Cell Biol. 2016, 214, 249–257. [Google Scholar] [CrossRef] [Green Version]
- Kory, N.; Wyant, G.A.; Prakash, G.; Uit de Bos, J.; Bottanelli, F.; Pacold, M.E.; Chan, S.H.; Lewis, C.A.; Wang, T.; Keys, H.R.; et al. SFXN1 is a mitochondrial serine transporter required for one-carbon metabolism. Science 2018, 362, eaat9528. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Zheng, J.; Shen, N.; Wang, G.; Zhou, G.; Fang, Y.; Lin, J.; Zhao, J. Identification of pathways and genes associated with synovitis in osteoarthritis using bioinformatics analyses. Sci. Rep. 2018, 8, 10050. [Google Scholar] [CrossRef] [Green Version]
- Li, H.-B.; Tong, J.; Zhu, S.; Batista, P.J.; Duffy, E.E.; Zhao, J.; Bailis, W.; Cao, G.; Kroehling, L.; Chen, Y.; et al. m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 2017, 548, 338–342. [Google Scholar] [CrossRef] [Green Version]
- Speakman, J.R. The ‘Fat Mass and Obesity Related’ (FTO) gene: Mechanisms of impact on obesity and energy balance. Curr. Obes. Rep. 2015, 4, 73–91. [Google Scholar] [CrossRef]
- Barbieri, I.; Kouzarides, T. Role of RNA modifications in cancer. Nat. Rev. Cancer 2020, 20, 303–322. [Google Scholar] [CrossRef]
- Shi, H.; Wei, J.; He, C. Where, when, and how: Context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 2019, 74, 640–650. [Google Scholar] [CrossRef]
- Eisenberg, E.; Levanon, E.Y. A-to-I RNA editing-immune protector and transcriptome diversifier. Nat. Rev. Genet. 2018, 19, 473–490. [Google Scholar] [CrossRef]
- Shulman, Z.; Stern-Ginossar, N. The RNA modification N6-methyladenosine as a novel regulator of the immune system. Nat. Immunol. 2020, 21, 501–512. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.; Feinbaum, R.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Lee, R.; Ambros, V. An extensive class of small RNAs in Caenorhabditis elegans. Science 2001, 294, 862–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, N.C.; Lim, L.P.; Weinstein, E.G.; Bartel, D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 2001, 294, 858–862. [Google Scholar] [CrossRef] [Green Version]
- Sioud, M. RNA interference: Story and mechanisms. Method. Mol. Biol. 2021, 2282, 1–15. [Google Scholar]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [Green Version]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Ortbauer, M.; Ripper, D.; Fuhrmann, T.; Lassi, M.; Auernigg-Haselmaier, S.; Stiegler, C.; König, J. Folate deficiency and over-supplementation causes impaired folate metabolism: Regulation and adaptation mechanisms in Caenorhabditis elegans. Mol. Nutr. Food Res. 2016, 60, 949–956. [Google Scholar] [CrossRef]
- Cabreiro, F.; Au, C.; Leung, K.Y.; Vergara-Irigaray, N.; Cochemé, H.M.; Noori, T.; Weinkove, D.; Schuster, E.; Greene, N.D.E.; Gems, D. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell 2013, 153, 228–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maynard, C.; Weinkove, D. The gut microbiota and ageing. Subcell. Biochem. 2018, 90, 351–371. [Google Scholar]
- García-González, A.P.; Ritter, A.D.; Shrestha, S.; Andersen, E.C.; Yilmaz, L.S.; Walhout, A.J.M. Bacterial metabolism affects the C. elegans response to cancer chemotherapeutics. Cell 2017, 169, 431–441.e8. [Google Scholar] [CrossRef] [Green Version]
- Ke, W.; Saba, J.A.; Yao, C.H.; Hilzendeger, M.A.; Drangowska-Way, A.; Joshi, C.; Mony, V.K.; Benjamin, S.B.; Zhang, S.; Locasale, J.; et al. Dietary serine-microbiota interaction enhances chemotherapeutic toxicity without altering drug conversion. Nat. Commun. 2020, 11, 2587. [Google Scholar] [CrossRef] [PubMed]
- Rosener, B.; Sayin, S.; Oluoch, P.O.; García González, A.P.; Mori, H.; Walhout, A.J.; Mitchell, A. Evolved bacterial resistance against fluoropyrimidines can lower chemotherapy impact in the Caenorhabditis elegans host. Elife 2020, 9, e59831. [Google Scholar] [CrossRef] [PubMed]
- Navarro, I.C.; Tuorto, F.; Jordan, D.; Legrand, C.; Price, J.; Braukmann, F.; Hendrick, A.G.; Akay, A.; Kotter, A.; Helm, M.; et al. Translational adaptation to heat stress is mediated by RNA 5-methylcytosine in Caenorhabditis elegans. EMBO J. 2021, 40, e105496. [Google Scholar] [CrossRef] [PubMed]
- Heissenberger, C.; Rollins, J.A.; Krammer, T.L.; Nagelreiter, F.; Stocker, I.; Wacheul, L.; Shpylovyi, A.; Tav, K.; Snow, S.; Grillari, J.; et al. The ribosomal RNA m5C methyltransferase NSUN-1 modulates healthspan and oogenesis in Caenorhabditis elegans. Elife 2020, 9, e56205. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takeda, Y.; Chijimatsu, R.; Vecchione, A.; Arai, T.; Kitagawa, T.; Ofusa, K.; Yabumoto, M.; Hirotsu, T.; Eguchi, H.; Doki, Y.; et al. Impact of One-Carbon Metabolism-Driving Epitranscriptome as a Therapeutic Target for Gastrointestinal Cancer. Int. J. Mol. Sci. 2021, 22, 7278. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22147278
Takeda Y, Chijimatsu R, Vecchione A, Arai T, Kitagawa T, Ofusa K, Yabumoto M, Hirotsu T, Eguchi H, Doki Y, et al. Impact of One-Carbon Metabolism-Driving Epitranscriptome as a Therapeutic Target for Gastrointestinal Cancer. International Journal of Molecular Sciences. 2021; 22(14):7278. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22147278
Chicago/Turabian StyleTakeda, Yu, Ryota Chijimatsu, Andrea Vecchione, Takahiro Arai, Toru Kitagawa, Ken Ofusa, Masami Yabumoto, Takaaki Hirotsu, Hidetoshi Eguchi, Yuichiro Doki, and et al. 2021. "Impact of One-Carbon Metabolism-Driving Epitranscriptome as a Therapeutic Target for Gastrointestinal Cancer" International Journal of Molecular Sciences 22, no. 14: 7278. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms22147278





